Targeting protein tyrosine phosphatases for anticancer drug discovery
- PMID: 20337577
- PMCID: PMC3076191
- DOI: 10.2174/138161210791209027
Targeting protein tyrosine phosphatases for anticancer drug discovery
Abstract
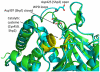
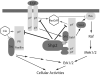
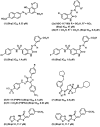
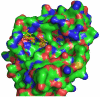
Protein tyrosine phosphatases (PTPs) are a diverse family of enzymes encoded by 107 genes in the human genome. Together with protein tyrosine kinases (PTKs), PTPs regulate various cellular activities essential for the initiation and maintenance of malignant phenotypes. While PTK inhibitors are now used routinely for cancer treatment, the PTP inhibitor development field is still in the discovery phase. In this article, the suitability of targeting PTPs for novel anticancer drug discovery is discussed. Examples are presented for PTPs that have been targeted for anticancer drug discovery as well as potential new PTP targets for novel anticancer drug discovery.
Figures
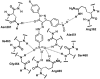

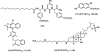
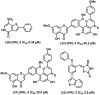

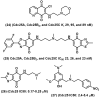
References
-
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. - PubMed
-
- Ostman A, Hellberg C, Bohmer FD. Protein-tyrosine phosphatases and cancer. Nat Rev Cancer. 2006;6:307–20. - PubMed
-
- Tonks NK. Protein tyrosine phosphatases: from genes, to function, to disease. Nat Rev Mol Cell Biol. 2006;7:833–46. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

