Metabolic modeling and analysis of the metabolic switch in Streptomyces coelicolor
- PMID: 20338070
- PMCID: PMC2853524
- DOI: 10.1186/1471-2164-11-202
Metabolic modeling and analysis of the metabolic switch in Streptomyces coelicolor
Abstract
Background: The transition from exponential to stationary phase in Streptomyces coelicolor is accompanied by a major metabolic switch and results in a strong activation of secondary metabolism. Here we have explored the underlying reorganization of the metabolome by combining computational predictions based on constraint-based modeling and detailed transcriptomics time course observations.
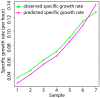
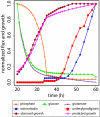
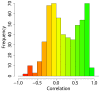
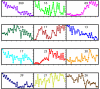
Results: We reconstructed the stoichiometric matrix of S. coelicolor, including the major antibiotic biosynthesis pathways, and performed flux balance analysis to predict flux changes that occur when the cell switches from biomass to antibiotic production. We defined the model input based on observed fermenter culture data and used a dynamically varying objective function to represent the metabolic switch. The predicted fluxes of many genes show highly significant correlation to the time series of the corresponding gene expression data. Individual mispredictions identify novel links between antibiotic production and primary metabolism.
Conclusion: Our results show the usefulness of constraint-based modeling for providing a detailed interpretation of time course gene expression data.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

