Mass action stoichiometric simulation models: incorporating kinetics and regulation into stoichiometric models
- PMID: 20338839
- PMCID: PMC2808481
- DOI: 10.1016/j.bpj.2009.09.064
Mass action stoichiometric simulation models: incorporating kinetics and regulation into stoichiometric models
Abstract
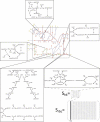
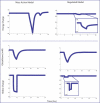
The ability to characterize biological dynamics is important for understanding the integrated molecular processes that underlie normal and abnormal cellular states. The availability of metabolomic data, in addition to new developments in the formal description of dynamic states of networks, has enabled a new data integration approach for building large-scale kinetic networks. We show that dynamic network models can be constructed in a scalable manner using metabolomic data mapped onto stoichiometric models, resulting in mass action stoichiometric simulation (MASS) models. Enzymes and their various functional states are represented explicitly as compounds, or nodes in a stoichiometric network, within this formalism. Analyses and simulations of MASS models explicitly show that regulatory enzymes can control dynamic states of networks in part by binding numerous metabolites at multiple sites. Thus, network functional states are reflected in the fractional states of a regulatory enzyme, such as the fraction of the total enzyme concentration that is in a catalytically active versus inactive state. The feasible construction of MASS models represents a practical means to increase the size, scope, and predictive capabilities of dynamic network models in cell and molecular biology.
Copyright 2010 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures



Similar articles
-
Towards kinetic modeling of genome-scale metabolic networks without sacrificing stoichiometric, thermodynamic and physiological constraints.Biotechnol J. 2013 Sep;8(9):1043-57. doi: 10.1002/biot.201300091. Epub 2013 Aug 20. Biotechnol J. 2013. PMID: 23868566
-
A unifying modelling formalism for the integration of stoichiometric and kinetic models.J R Soc Interface. 2020 Aug;17(169):20200341. doi: 10.1098/rsif.2020.0341. Epub 2020 Aug 5. J R Soc Interface. 2020. PMID: 32752999 Free PMC article.
-
MASSpy: Building, simulating, and visualizing dynamic biological models in Python using mass action kinetics.PLoS Comput Biol. 2021 Jan 28;17(1):e1008208. doi: 10.1371/journal.pcbi.1008208. eCollection 2021 Jan. PLoS Comput Biol. 2021. PMID: 33507922 Free PMC article.
-
Modeling metabolic networks for mammalian cell systems: general considerations, modeling strategies, and available tools.Adv Biochem Eng Biotechnol. 2012;127:71-108. doi: 10.1007/10_2011_120. Adv Biochem Eng Biotechnol. 2012. PMID: 21984615 Review.
-
Comparing methods for metabolic network analysis and an application to metabolic engineering.Gene. 2013 May 25;521(1):1-14. doi: 10.1016/j.gene.2013.03.017. Epub 2013 Mar 26. Gene. 2013. PMID: 23537990 Review.
Cited by
-
Systems-based approaches to study immunometabolism.Cell Mol Immunol. 2022 Mar;19(3):409-420. doi: 10.1038/s41423-021-00783-9. Epub 2022 Feb 4. Cell Mol Immunol. 2022. PMID: 35121805 Free PMC article. Review.
-
k-OptForce: integrating kinetics with flux balance analysis for strain design.PLoS Comput Biol. 2014 Feb 20;10(2):e1003487. doi: 10.1371/journal.pcbi.1003487. eCollection 2014 Feb. PLoS Comput Biol. 2014. PMID: 24586136 Free PMC article.
-
A cooperative strategy for parameter estimation in large scale systems biology models.BMC Syst Biol. 2012 Jun 22;6:75. doi: 10.1186/1752-0509-6-75. BMC Syst Biol. 2012. PMID: 22727112 Free PMC article.
-
Community-scale models of microbiomes: Articulating metabolic modelling and metagenome sequencing.Microb Biotechnol. 2024 Jan;17(1):e14396. doi: 10.1111/1751-7915.14396. Epub 2024 Jan 20. Microb Biotechnol. 2024. PMID: 38243750 Free PMC article. Review.
-
The development of a fully-integrated immune response model (FIRM) simulator of the immune response through integration of multiple subset models.BMC Syst Biol. 2013 Sep 28;7:95. doi: 10.1186/1752-0509-7-95. BMC Syst Biol. 2013. PMID: 24074340 Free PMC article.
References
-
- Scriver C. McGraw-Hill Professional; New York: 2000. The Metabolic and Molecular Bases of Inherited Disease.
-
- Teusink B., Passarge J., Snoep J.L. Can yeast glycolysis be understood in terms of in vitro kinetics of the constituent enzymes? Testing biochemistry. Eur. J. Biochem. 2000;267:5313–5329. - PubMed
-
- Joshi A., Palsson B.O. Metabolic dynamics in the human red cell. Part III—Metabolic reaction rates. J. Theor. Biol. 1990;142:41–68. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

