A functional human Poly(A) site requires only a potent DSE and an A-rich upstream sequence
- PMID: 20339349
- PMCID: PMC2876958
- DOI: 10.1038/emboj.2010.42
A functional human Poly(A) site requires only a potent DSE and an A-rich upstream sequence
Abstract
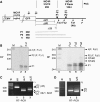
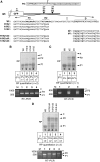
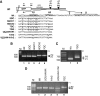
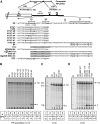
We have analysed the sequences required for cleavage and polyadenylation in the intronless melanocortin 4 receptor (MC4R) pre-mRNA. Unlike other intronless genes, 3'end processing of the MC4R primary transcript is independent of any auxiliary sequence elements and only requires the core poly(A) sequences. Mutation of the AUUAAA hexamer had little effect on MC4R 3'end processing but small changes in the short DSE severely reduced cleavage efficiency. The MC4R poly(A) site requires only the DSE and an A-rich upstream sequence to direct efficient cleavage and polyadenylation. Our observation may be highly relevant for the understanding of how human noncanonical poly(A) sites are recognised. This is supported by a genome-wide analysis of over 10 000 poly(A) sites where we show that many human noncanonical poly(A) signals contain A-rich upstream sequences and tend to have a higher frequency of U and GU nucleotides in their DSE compared with canonical poly(A) signals. The importance of A-rich elements for noncanonical poly(A) site recognition was confirmed by mutational analysis of the human JUNB gene, which contains an A-rich noncanonical poly(A) signal.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

Comment in
-
Polyadenylation: alternative lifestyles of the A-rich (and famous?).EMBO J. 2010 May 5;29(9):1473-4. doi: 10.1038/emboj.2010.67. EMBO J. 2010. PMID: 20442727 Free PMC article. No abstract available.
References
-
- Anand S, Batista FD, Tkach T, Efremov DG, Burrone OR (1997) Multiple transcripts of the murine immunoglobulin epsilon membrane locus are generated by alternative splicing and differential usage of two polyadenylation sites. Mol Immunol 34: 175–183 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

