Cytoplasmic polyadenylation element binding protein is a conserved target of tumor suppressor HRPT2/CDC73
- PMID: 20339377
- PMCID: PMC3109426
- DOI: 10.1038/cdd.2010.32
Cytoplasmic polyadenylation element binding protein is a conserved target of tumor suppressor HRPT2/CDC73
Abstract
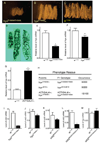
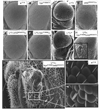
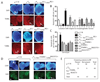
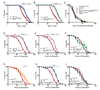
Parafibromin, a tumor suppressor protein encoded by HRPT2/CDC73 and implicated in parathyroid cancer and the hyperparathyroidism-jaw tumor (HPT-JT) familial cancer syndrome, is part of the PAF1 transcriptional regulatory complex. Parafibromin has been implicated in apoptosis and growth arrest, but the mechanism by which its loss of function promotes neoplasia is poorly understood. In this study we report that a hypomorphic allele of hyrax (hyx), the Drosophila homolog of HRPT2/CDC73, rescues the loss-of-ventral-eye phenotype of lobe (Akt1s1). Such rescue is consistent with previous reports that hyx/parafibromin is required for the nuclear transduction of Wingless (Wg)/Wnt signals and that Wg signaling antagonizes lobe function. A screen using double hyx/lobe heterozygotes identified an additional interaction with orb and orb2, the homologs of mammalian cytoplasmic polyadenylation element binding protein (CPEB), a translational regulatory protein. Hyx and orb2 heterozygotes lived longer and were more resistant to starvation than controls. In mammalian cells, knockdown of parafibromin expression reduced levels of CPEB1. Chromatin immunoprecipitation (ChIP) showed occupancy of CPEB1 by endogenous parafibromin. Bioinformatic analysis revealed a significant overlap between human transcripts potentially regulated by parafibromin and CPEB. These results show that parafibromin may exert both transcriptional and, through CPEB, translational control over a subset of target genes and that loss of parafibromin (and CPEB) function may promote tumorigenesis in part by conferring resistance to nutritional stress.
Conflict of interest statement
None of the authors has a conflict of interest that could be perceived as prejudicing the impartiality of the research reported herein.
Figures


Similar articles
-
The EIF4EBP3 translational repressor is a marker of CDC73 tumor suppressor haploinsufficiency in a parathyroid cancer syndrome.Cell Death Dis. 2012 Feb 2;3(2):266. doi: 10.1038/cddis.2012.6. Cell Death Dis. 2012. PMID: 22297294 Free PMC article.
-
The role of Parafibromin/Hyrax as a nuclear Gli/Ci-interacting protein in Hedgehog target gene control.Mech Dev. 2009 May-Jun;126(5-6):394-405. doi: 10.1016/j.mod.2009.02.002. Epub 2009 Feb 11. Mech Dev. 2009. PMID: 19368795
-
Defective nucleolar localization and dominant interfering properties of a parafibromin L95P missense mutant causing the hyperparathyroidism-jaw tumor syndrome.Endocr Relat Cancer. 2010 May 18;17(2):513-24. doi: 10.1677/ERC-09-0272. Print 2010 Jun. Endocr Relat Cancer. 2010. PMID: 20304979 Free PMC article.
-
The roles of the tumor suppressor parafibromin in cancer.Front Cell Dev Biol. 2022 Sep 21;10:1006400. doi: 10.3389/fcell.2022.1006400. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36211470 Free PMC article. Review.
-
Tumor suppressor gene mutation in a patient with a history of hyperparathyroidism-jaw tumor syndrome and healed generalized osteitis fibrosa cystica: a case report and genetic pathophysiology review.J Oral Maxillofac Surg. 2015 Jan;73(1):194.e1-9. doi: 10.1016/j.joms.2014.09.008. Epub 2014 Sep 28. J Oral Maxillofac Surg. 2015. PMID: 25511968 Review.
Cited by
-
The many roles of the conserved eukaryotic Paf1 complex in regulating transcription, histone modifications, and disease states.Biochim Biophys Acta. 2013 Jan;1829(1):116-26. doi: 10.1016/j.bbagrm.2012.08.011. Epub 2012 Sep 6. Biochim Biophys Acta. 2013. PMID: 22982193 Free PMC article.
-
CPEB1 promotes differentiation and suppresses EMT in mammary epithelial cells.J Cell Sci. 2014 May 15;127(Pt 10):2326-38. doi: 10.1242/jcs.144956. Epub 2014 Mar 14. J Cell Sci. 2014. PMID: 24634508 Free PMC article.
-
The EIF4EBP3 translational repressor is a marker of CDC73 tumor suppressor haploinsufficiency in a parathyroid cancer syndrome.Cell Death Dis. 2012 Feb 2;3(2):266. doi: 10.1038/cddis.2012.6. Cell Death Dis. 2012. PMID: 22297294 Free PMC article.
-
Cataloguing and Selection of mRNAs Localized to Dendrites in Neurons and Regulated by RNA-Binding Proteins in RNA Granules.Biomolecules. 2020 Jan 22;10(2):167. doi: 10.3390/biom10020167. Biomolecules. 2020. PMID: 31978946 Free PMC article. Review.
-
The mitochondrial calcium uniporter contributes to morphine tolerance through pCREB and CPEB1 in rat spinal cord dorsal horn.Br J Anaesth. 2019 Aug;123(2):e226-e238. doi: 10.1016/j.bja.2019.05.027. Epub 2019 Jun 26. Br J Anaesth. 2019. PMID: 31253357 Free PMC article.
References
-
- Jackson CE, Norum RA, Boyd SB, Talpos GB, Wilson SD, Taggart RT, et al. Hereditary hyperparathyroidism and multiple ossifying jaw fibromas: a clinically and genetically distinct syndrome. Surgery. 1990;108:1006–1012. - PubMed
-
- Mallette LE, Malini S, Rappaport MP, Kirkland JL. Familial cystic parathyroid adenomatosis. Ann Intern Med. 1987;107:54–60. - PubMed
-
- Teh BT, Farnebo F, Kristoffersson U, Sundelin B, Cardinal J, Axelson R, et al. Autosomal dominant primary hyperparathyroidism and jaw tumor syndrome associated with renal hamartomas and cystic kidney disease: linkage to 1q21-q32 and loss of the wild type allele in renal hamartomas. J Clin Endocrinol Metab. 1996;81:4204–4211. - PubMed
-
- Teh BT, Farnebo F, Twigg S, Höög A, Kytölä S, Korpi-Hyövälti E, et al. Familial isolated hyperparathyroidism maps to the hyperparathyroidism-jaw tumor locus in 1q21-q32 in a subset of families. J Clin Endocrinol Metab. 1998;83:2114–2120. - PubMed
-
- Simonds WF, James-Newton LA, Agarwal SK, Yang B, Skarulis MC, Hendy GN, et al. Familial isolated hyperparathyroidism: Clinical and genetic characteristics of thirty-six kindreds. Medicine (Baltimore) 2002;81:1–26. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

