The short-chain oxidoreductase Q9HYA2 from Pseudomonas aeruginosa PAO1 contains an atypical catalytic center
- PMID: 20340135
- PMCID: PMC2868251
- DOI: 10.1002/pro.384
The short-chain oxidoreductase Q9HYA2 from Pseudomonas aeruginosa PAO1 contains an atypical catalytic center
Abstract
The characteristic oxidation or reduction reaction mechanisms of short-chain oxidoreductase (SCOR) enzymes involve a highly conserved Asp-Ser-Tyr-Lys catalytic tetrad. The SCOR enzyme Q9HYA2 from the pathogenic bacterium Pseudomonas aeruginosa was recognized to possess an atypical catalytic tetrad composed of Lys118-Ser146-Thr159-Arg163. Orthologs of Q9HYA2 containing the unusual catalytic tetrad along with conserved substrate and cofactor recognition residues were identified in 27 additional species, the majority of which are bacterial pathogens. However, this atypical catalytic tetrad was not represented within the Protein Data Bank. The crystal structures of unligated and NADPH-complexed Q9HYA2 were determined at 2.3 A resolution. Structural alignment to a polyketide ketoreductase (KR), a typical SCOR, demonstrated that Q9HYA2's Lys118, Ser146, and Arg163 superimposed upon the KR's catalytic Asp114, Ser144, and Lys161, respectively. However, only the backbone of Q9HYA2's Thr159 overlapped KR's catalytic Tyr157. The Thr159 hydroxyl in apo Q9HYA2 is poorly positioned for participating in catalysis. In the Q9HYA2-NADPH complex, the Thr159 side chain was modeled in two alternate rotamers, one of which is positioned to interact with other members of the tetrad and the bound cofactor. A chloride ion is bound at the position normally occupied by the catalytic tyrosine hydroxyl. The putative active site of Q9HYA2 contains a chemical moiety at each catalytically important position of a typical SCOR enzyme. This is the first observation of a SCOR protein with this alternate catalytic center that includes threonine replacing the catalytic tyrosine and an ion replacing the hydroxyl moiety of the catalytic tyrosine.
Figures
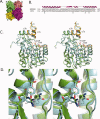
Similar articles
-
Structural analysis of actinorhodin polyketide ketoreductase: cofactor binding and substrate specificity.Biochemistry. 2004 Nov 23;43(46):14529-38. doi: 10.1021/bi048133a. Biochemistry. 2004. PMID: 15544323 Free PMC article.
-
Rational proteomics I. Fingerprint identification and cofactor specificity in the short-chain oxidoreductase (SCOR) enzyme family.Proteins. 2003 Dec 1;53(4):931-43. doi: 10.1002/prot.10512. Proteins. 2003. PMID: 14635134
-
Substrate Channel Flexibility in Pseudomonas aeruginosa MurB Accommodates Two Distinct Substrates.PLoS One. 2013 Jun 21;8(6):e66936. doi: 10.1371/journal.pone.0066936. Print 2013. PLoS One. 2013. PMID: 23805286 Free PMC article.
-
Rational proteomics II: electrostatic nature of cofactor preference in the short-chain oxidoreductase (SCOR) enzyme family.Proteins. 2004 Nov 1;57(2):294-301. doi: 10.1002/prot.20205. Proteins. 2004. PMID: 15340916 Free PMC article.
-
Medium- and short-chain dehydrogenase/reductase gene and protein families : the SDR superfamily: functional and structural diversity within a family of metabolic and regulatory enzymes.Cell Mol Life Sci. 2008 Dec;65(24):3895-906. doi: 10.1007/s00018-008-8588-y. Cell Mol Life Sci. 2008. PMID: 19011750 Free PMC article. Review.
Cited by
-
SDR7-6, a short-chain alcohol dehydrogenase/reductase family protein, regulates light-dependent cell death and defence responses in rice.Mol Plant Pathol. 2022 Jan;23(1):78-91. doi: 10.1111/mpp.13144. Epub 2021 Oct 11. Mol Plant Pathol. 2022. PMID: 34633131 Free PMC article.
References
-
- Tanaka N, Nonaka T, Nakamura KT, Hara A. SDR structure, mechanism of action, and substrate recognition. Curr Org Chem. 2001;5:89–111.
-
- Duax WL, Pletnev V, Addlagatta A, Bruenn J, Weeks CM. Rational proteomics. I. Fingerprint identification and cofactor specificity in the short-chain oxidoreductase (SCOR) enzyme family. Proteins. 2003;53:931–943. - PubMed
-
- Rossolini GM, Mantengoli E. Treatment and control of severe infections caused by multiresistant Pseudomonas aeruginosa. Clin Microbiol Infect. 2005;11(Suppl 4):17–32. - PubMed
-
- Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis. 2009;48:1–12. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

