Physical mapping in highly heterozygous genomes: a physical contig map of the Pinot Noir grapevine cultivar
- PMID: 20346114
- PMCID: PMC2865496
- DOI: 10.1186/1471-2164-11-204
Physical mapping in highly heterozygous genomes: a physical contig map of the Pinot Noir grapevine cultivar
Abstract
Background: Most of the grapevine (Vitis vinifera L.) cultivars grown today are those selected centuries ago, even though grapevine is one of the most important fruit crops in the world. Grapevine has therefore not benefited from the advances in modern plant breeding nor more recently from those in molecular genetics and genomics: genes controlling important agronomic traits are practically unknown. A physical map is essential to positionally clone such genes and instrumental in a genome sequencing project.
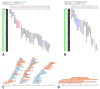
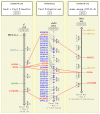
Results: We report on the first whole genome physical map of grapevine built using high information content fingerprinting of 49,104 BAC clones from the cultivar Pinot Noir. Pinot Noir, as most grape varieties, is highly heterozygous at the sequence level. This resulted in the two allelic haplotypes sometimes assembling into separate contigs that had to be accommodated in the map framework or in local expansions of contig maps. We performed computer simulations to assess the effects of increasing levels of sequence heterozygosity on BAC fingerprint assembly and showed that the experimental assembly results are in full agreement with the theoretical expectations, given the heterozygosity levels reported for grape. The map is anchored to a dense linkage map consisting of 994 markers. 436 contigs are anchored to the genetic map, covering 342 of the 475 Mb that make up the grape haploid genome.
Conclusions: We have developed a resource that makes it possible to access the grapevine genome, opening the way to a new era both in grape genetics and breeding and in wine making. The effects of heterozygosity on the assembly have been analyzed and characterized by using several complementary approaches which could be easily transferred to the study of other genomes which present the same features.
Figures
Similar articles
-
A dense single-nucleotide polymorphism-based genetic linkage map of grapevine (Vitis vinifera L.) anchoring Pinot Noir bacterial artificial chromosome contigs.Genetics. 2007 Aug;176(4):2637-50. doi: 10.1534/genetics.106.067462. Epub 2007 Jul 1. Genetics. 2007. PMID: 17603124 Free PMC article.
-
A physical map of the heterozygous grapevine 'Cabernet Sauvignon' allows mapping candidate genes for disease resistance.BMC Plant Biol. 2008 Jun 13;8:66. doi: 10.1186/1471-2229-8-66. BMC Plant Biol. 2008. PMID: 18554400 Free PMC article.
-
A high quality draft consensus sequence of the genome of a heterozygous grapevine variety.PLoS One. 2007 Dec 19;2(12):e1326. doi: 10.1371/journal.pone.0001326. PLoS One. 2007. PMID: 18094749 Free PMC article.
-
Resurgence of minority and autochthonous grapevine varieties in South America: a review of their oenological potential.J Sci Food Agric. 2020 Jan 30;100(2):465-482. doi: 10.1002/jsfa.10003. Epub 2019 Oct 29. J Sci Food Agric. 2020. PMID: 31452209 Review.
-
A review on the aroma composition of Vitis vinifera L. Pinot noir wines: origins and influencing factors.Crit Rev Food Sci Nutr. 2021;61(10):1589-1604. doi: 10.1080/10408398.2020.1762535. Epub 2020 May 13. Crit Rev Food Sci Nutr. 2021. PMID: 32401040 Review.
Cited by
-
Facing Climate Change: Biotechnology of Iconic Mediterranean Woody Crops.Front Plant Sci. 2019 Apr 16;10:427. doi: 10.3389/fpls.2019.00427. eCollection 2019. Front Plant Sci. 2019. PMID: 31057569 Free PMC article. Review.
-
Identification of tissue-specific, abiotic stress-responsive gene expression patterns in wine grape (Vitis vinifera L.) based on curation and mining of large-scale EST data sets.BMC Plant Biol. 2011 May 18;11:86. doi: 10.1186/1471-2229-11-86. BMC Plant Biol. 2011. PMID: 21592389 Free PMC article.
-
A framework genetic map of Muscadinia rotundifolia.Theor Appl Genet. 2012 Oct;125(6):1195-210. doi: 10.1007/s00122-012-1906-7. Epub 2012 Jun 12. Theor Appl Genet. 2012. PMID: 22688272
-
Physical mapping and BAC-end sequence analysis provide initial insights into the flax (Linum usitatissimum L.) genome.BMC Genomics. 2011 May 9;12:217. doi: 10.1186/1471-2164-12-217. BMC Genomics. 2011. PMID: 21554714 Free PMC article.
-
Advances in biotechnology and informatics to link variation in the genome to phenotypes in plants and animals.Funct Integr Genomics. 2013 Mar;13(1):1-9. doi: 10.1007/s10142-013-0319-2. Epub 2013 Mar 15. Funct Integr Genomics. 2013. PMID: 23494190 Free PMC article. Review.
References
-
- Salmaso M, Faes G, Segala C, Stefanini M, Salakhutdinov I, Zyprian E, Toepfer R, Grando M, Velasco R. Genome diversity and gene haplotypes in the grapevine (Vitis vinifera L.), as revealed by single nucleotide polymorphisms. Mol Breed. 2005;14:385–395. doi: 10.1007/s11032-005-0261-7. - DOI
-
- Lamoureux D, Bernole A, Le Clainche I, Tual S, Thareau V, Paillard S, Legeai F, Dossat C, Wincker P, Oswald M. et al.Anchoring of a large set of markers onto a BAC library for the development of a draft physical map of the grapevine genome. Theor Appl Genet. 2006;113:344–356. doi: 10.1007/s00122-006-0301-7. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

