Effects of varying Notch1 signal strength on embryogenesis and vasculogenesis in compound mutant heterozygotes
- PMID: 20346184
- PMCID: PMC2865454
- DOI: 10.1186/1471-213X-10-36
Effects of varying Notch1 signal strength on embryogenesis and vasculogenesis in compound mutant heterozygotes
Abstract
Background: Identifying developmental processes regulated by Notch1 can be addressed in part by characterizing mice with graded levels of Notch1 signaling strength. Here we examine development in embryos expressing various combinations of Notch1 mutant alleles. Mice homozygous for the hypomorphic Notch1(12f) allele, which removes the single O-fucose glycan in epidermal growth factor-like repeat 12 (EGF12) of the Notch1 ligand binding domain (lbd), exhibit reduced growth after weaning and defective T cell development. Mice homozygous for the inactive Notch1(lbd) allele express Notch1 missing an aproximately 20 kDa internal segment including the canonical Notch1 ligand binding domain, and die at embryonic day approximately E9.5. The embryonic and vascular phenotypes of compound heterozygous Notch1(12f/lbd) embryos were compared with Notch1+/12f, Notch1(12f/12f), and Notch1(lbd/lbd) embryos. Embryonic stem (ES) cells derived from these embryos were also examined in Notch signaling assays. While Notch1 signaling was stronger in Notch1(12f/lbd) compound heterozygotes compared to Notch1(lbd/lbd) embryos and ES cells, Notch1 signaling was even stronger in embryos carrying Notch1(12f) and a null Notch1 allele.
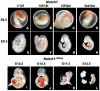
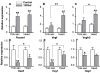
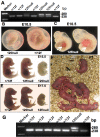
Results: Mouse embryos expressing the hypomorphic Notch1(12f) allele, in combination with the inactive Notch1(lbd) allele which lacks the Notch1 ligand binding domain, died at approximately E11.5-12.5. Notch1(12f/lbd) ES cells signaled less well than Notch1(12f/12f) ES cells but more strongly than Notch1lbd/lbd ES cells. However, vascular defects in Notch1(12f/lbd) yolk sac were severe and similar to Notch1(lbd/lbd) yolk sac. By contrast, vascular disorganization was milder in Notch1(12f/lbd) compared to Notch1(lbd/lbd) embryos. The expression of Notch1 target genes was low in Notch1(12f/lbd) yolk sac and embryo head, whereas Vegf and Vegfr2 transcripts were increased. The severity of the compound heterozygous Notch1(12f/lbd) yolk sac phenotype suggested that the allelic products may functionally interact. By contrast, compound heterozygotes with Notch112f in combination with a Notch1 null allele (Notch1(tm1Con)) were capable of surviving to birth.
Conclusions: Notch1 signaling in Notch1(12f/lbd) compound heterozygous embryos is more defective than in compound heterozygotes expressing a hypomorphic Notch1(12f) allele and a Notch1 null allele. The data suggest that the gene products Notch1(lbd) and Notch1(12f) interact to reduce the activity of Notch1(12f).
Figures


Similar articles
-
A modifier in the 129S2/SvPasCrl genome is responsible for the viability of Notch1[12f/12f] mice.BMC Dev Biol. 2019 Oct 7;19(1):19. doi: 10.1186/s12861-019-0199-3. BMC Dev Biol. 2019. PMID: 31590629 Free PMC article.
-
The O-fucose glycan in the ligand-binding domain of Notch1 regulates embryogenesis and T cell development.Proc Natl Acad Sci U S A. 2008 Feb 5;105(5):1539-44. doi: 10.1073/pnas.0702846105. Epub 2008 Jan 28. Proc Natl Acad Sci U S A. 2008. PMID: 18227520 Free PMC article.
-
In vivo consequences of deleting EGF repeats 8-12 including the ligand binding domain of mouse Notch1.BMC Dev Biol. 2008 Apr 29;8:48. doi: 10.1186/1471-213X-8-48. BMC Dev Biol. 2008. PMID: 18445292 Free PMC article.
-
Analysis of homozygous TGF beta 1 null mouse embryos demonstrates defects in yolk sac vasculogenesis and hematopoiesis.Ann N Y Acad Sci. 1995 Mar 27;752:300-8. doi: 10.1111/j.1749-6632.1995.tb17439.x. Ann N Y Acad Sci. 1995. PMID: 7755275 Review. No abstract available.
-
Self-organized signaling in stem cell models of embryos.Stem Cell Reports. 2021 May 11;16(5):1065-1077. doi: 10.1016/j.stemcr.2021.03.020. Stem Cell Reports. 2021. PMID: 33979594 Free PMC article. Review.
Cited by
-
Protein O-fucosyltransferase 1 (Pofut1) regulates lymphoid and myeloid homeostasis through modulation of Notch receptor ligand interactions.Blood. 2011 May 26;117(21):5652-62. doi: 10.1182/blood-2010-12-326074. Epub 2011 Apr 4. Blood. 2011. PMID: 21464368 Free PMC article.
-
O-GlcNAc on NOTCH1 EGF repeats regulates ligand-induced Notch signaling and vascular development in mammals.Elife. 2017 Apr 11;6:e24419. doi: 10.7554/eLife.24419. Elife. 2017. PMID: 28395734 Free PMC article.
-
The mouse allantois: new insights at the embryonic-extraembryonic interface.Philos Trans R Soc Lond B Biol Sci. 2022 Dec 5;377(1865):20210251. doi: 10.1098/rstb.2021.0251. Epub 2022 Oct 17. Philos Trans R Soc Lond B Biol Sci. 2022. PMID: 36252214 Free PMC article. Review.
-
Notch post-translationally regulates β-catenin protein in stem and progenitor cells.Nat Cell Biol. 2011 Aug 14;13(10):1244-51. doi: 10.1038/ncb2313. Nat Cell Biol. 2011. PMID: 21841793 Free PMC article.
-
What Have We Learned from Glycosyltransferase Knockouts in Mice?J Mol Biol. 2016 Aug 14;428(16):3166-3182. doi: 10.1016/j.jmb.2016.03.025. Epub 2016 Mar 31. J Mol Biol. 2016. PMID: 27040397 Free PMC article. Review.
References
-
- Schweisguth F. Regulation of notch signaling activity. Curr Biol. 2004;14:R129–38. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

