A functional link between the histone demethylase PHF8 and the transcription factor ZNF711 in X-linked mental retardation
- PMID: 20346720
- PMCID: PMC2989439
- DOI: 10.1016/j.molcel.2010.03.002
A functional link between the histone demethylase PHF8 and the transcription factor ZNF711 in X-linked mental retardation
Abstract
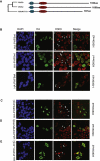
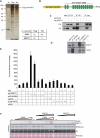
X-linked mental retardation (XLMR) is an inherited disorder that mostly affects males and is caused by mutations in genes located on the X chromosome. Here, we show that the XLMR protein PHF8 and a C. elegans homolog F29B9.2 catalyze demethylation of di- and monomethylated lysine 9 of histone H3 (H3K9me2/me1). The PHD domain of PHF8 binds to H3K4me3 and colocalizes with H3K4me3 at transcription initiation sites. Furthermore, PHF8 interacts with another XMLR protein, ZNF711, which binds to a subset of PHF8 target genes, including the XLMR gene JARID1C. Of interest, the C. elegans PHF8 homolog is highly expressed in neurons, and mutant animals show impaired locomotion. Taken together, our results functionally link the XLMR gene PHF8 to two other XLMR genes, ZNF711 and JARID1C, indicating that MR genes may be functionally linked in pathways, causing the complex phenotypes observed in patients developing MR.
Copyright 2010 Elsevier Inc. All rights reserved.
Figures





Comment in
-
Demethylases go mental.Mol Cell. 2010 Apr 23;38(2):155-7. doi: 10.1016/j.molcel.2010.04.002. Mol Cell. 2010. PMID: 20417593 No abstract available.
Similar articles
-
The X-linked mental retardation gene PHF8 is a histone demethylase involved in neuronal differentiation.Cell Res. 2010 Aug;20(8):908-18. doi: 10.1038/cr.2010.81. Epub 2010 Jun 15. Cell Res. 2010. PMID: 20548336
-
Histone H4K20/H3K9 demethylase PHF8 regulates zebrafish brain and craniofacial development.Nature. 2010 Jul 22;466(7305):503-7. doi: 10.1038/nature09261. Epub 2010 Jul 11. Nature. 2010. PMID: 20622853 Free PMC article.
-
The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases.Cell. 2007 Mar 23;128(6):1077-88. doi: 10.1016/j.cell.2007.02.017. Epub 2007 Feb 22. Cell. 2007. PMID: 17320160
-
X-linked mental retardation: further lumping, splitting and emerging phenotypes.Clin Genet. 2005 Jun;67(6):451-67. doi: 10.1111/j.1399-0004.2005.00434.x. Clin Genet. 2005. PMID: 15857409 Review.
-
X-linked mental retardation.Med Sci Monit. 2008 Nov;14(11):RA221-9. Med Sci Monit. 2008. PMID: 18971887 Review.
Cited by
-
Cyclin E-CDK2 protein phosphorylates plant homeodomain finger protein 8 (PHF8) and regulates its function in the cell cycle.J Biol Chem. 2015 Feb 13;290(7):4075-85. doi: 10.1074/jbc.M114.602532. Epub 2014 Dec 29. J Biol Chem. 2015. PMID: 25548279 Free PMC article.
-
Identification and characterization of nardilysin as a novel dimethyl H3K4-binding protein involved in transcriptional regulation.J Biol Chem. 2012 Mar 23;287(13):10089-10098. doi: 10.1074/jbc.M111.313965. Epub 2012 Jan 30. J Biol Chem. 2012. PMID: 22294699 Free PMC article.
-
Increasing Notch signaling antagonizes PRC2-mediated silencing to promote reprograming of germ cells into neurons.Elife. 2016 Sep 7;5:e15477. doi: 10.7554/eLife.15477. Elife. 2016. PMID: 27602485 Free PMC article.
-
PHF8 Plays an Oncogene Function in Hepatocellular Carcinoma Formation.Oncol Res. 2019 May 7;27(5):613-621. doi: 10.3727/096504018X15410353669149. Epub 2019 Feb 14. Oncol Res. 2019. PMID: 30764899 Free PMC article.
-
The G2/M regulator histone demethylase PHF8 is targeted for degradation by the anaphase-promoting complex containing CDC20.Mol Cell Biol. 2013 Nov;33(21):4166-80. doi: 10.1128/MCB.00689-13. Epub 2013 Aug 26. Mol Cell Biol. 2013. PMID: 23979597 Free PMC article.
References
-
- American Psychiatric Association . Fourth Edition. The American Psychiatric Association; Washington, DC: 1994. Diagnostic and statistical manual of mental disorders.
-
- Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

