The histone chaperone Nap1 promotes nucleosome assembly by eliminating nonnucleosomal histone DNA interactions
- PMID: 20347425
- PMCID: PMC2880918
- DOI: 10.1016/j.molcel.2010.01.037
The histone chaperone Nap1 promotes nucleosome assembly by eliminating nonnucleosomal histone DNA interactions
Abstract
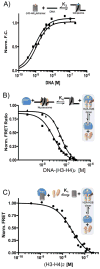
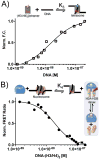
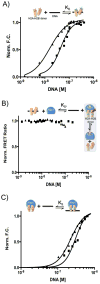
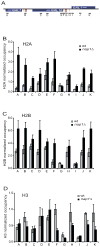
The organization of the eukaryotic genome into nucleosomes dramatically affects the regulation of gene expression. The delicate balance between transcription and DNA compaction relies heavily on nucleosome dynamics. Surprisingly, little is known about the free energy required to assemble these large macromolecular complexes and maintain them under physiological conditions. Here, we describe the thermodynamic parameters that drive nucleosome formation in vitro. To demonstrate the versatility of our approach, we test the effect of DNA sequence and H3K56 acetylation on nucleosome thermodynamics. Furthermore, our studies reveal the mechanism of action of the histone chaperone nucleosome assembly protein 1 (Nap1). We present evidence for a paradigm in which nucleosome assembly requires the elimination of competing, nonnucleosomal histone-DNA interactions by Nap1. This observation is confirmed in vivo, wherein deletion of the NAP1 gene in yeast results in a significant increase in atypical histone-DNA complexes, as well as in deregulated transcription activation and repression.
(c) 2010 Elsevier Inc. All rights reserved.
Figures

Comment in
-
Avoiding a fatal attraction: properties of nucleosomes and a histone chaperone revealed under physiological conditions.Mol Cell. 2010 Mar 26;37(6):747-8. doi: 10.1016/j.molcel.2010.02.016. Mol Cell. 2010. PMID: 20347417
References
-
- Akey CW, Luger K. Histone chaperones and nucleosome assembly. Curr Opin Struct Biol. 2003;13:6–14. - PubMed
-
- Aragay AM, Diaz P, Daban JR. Association of nucleosome core particle DNA with different histone oligomers. Transfer of histones between DNA-(H2A, H2B) and DNA-(H3, H4) complexes. J Mol Biol. 1988;204:141–154. - PubMed
-
- Bryant GO, Ptashne M. Independent recruitment in vivo by Gal4 of two complexes required for transcription. Mol Cell. 2003;11:1301–1309. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

