Oxidative activity of yeast Ero1p on protein disulfide isomerase and related oxidoreductases of the endoplasmic reticulum
- PMID: 20348090
- PMCID: PMC2881739
- DOI: 10.1074/jbc.M109.064931
Oxidative activity of yeast Ero1p on protein disulfide isomerase and related oxidoreductases of the endoplasmic reticulum
Abstract
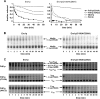
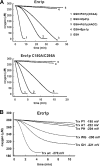
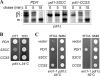
The sulfhydryl oxidase Ero1 oxidizes protein disulfide isomerase (PDI), which in turn catalyzes disulfide formation in proteins folding in the endoplasmic reticulum (ER). The extent to which other members of the PDI family are oxidized by Ero1 and thus contribute to net disulfide formation in the ER has been an open question. The yeast ER contains four PDI family proteins with at least one potential redox-active cysteine pair. We monitored the direct oxidation of each redox-active site in these proteins by yeast Ero1p in vitro. In this study, we found that the Pdi1p amino-terminal domain was oxidized most rapidly compared with the other oxidoreductase active sites tested, including the Pdi1p carboxyl-terminal domain. This observation is consistent with experiments conducted in yeast cells. In particular, the amino-terminal domain of Pdi1p preferentially formed mixed disulfides with Ero1p in vivo, and we observed synthetic lethality between a temperature-sensitive Ero1p variant and mutant Pdi1p lacking the amino-terminal active-site disulfide. Thus, the amino-terminal domain of yeast Pdi1p is on a preferred pathway for oxidizing the ER thiol pool. Overall, our results provide a rank order for the tendency of yeast ER oxidoreductases to acquire disulfides from Ero1p.
Figures





Similar articles
-
Balanced Ero1 activation and inactivation establishes ER redox homeostasis.J Cell Biol. 2012 Mar 19;196(6):713-25. doi: 10.1083/jcb.201110090. Epub 2012 Mar 12. J Cell Biol. 2012. PMID: 22412017 Free PMC article.
-
Novel Roles of the Non-catalytic Elements of Yeast Protein-disulfide Isomerase in Its Interplay with Endoplasmic Reticulum Oxidoreductin 1.J Biol Chem. 2016 Apr 8;291(15):8283-94. doi: 10.1074/jbc.M115.694257. Epub 2016 Feb 4. J Biol Chem. 2016. PMID: 26846856 Free PMC article.
-
Ero1p oxidizes protein disulfide isomerase in a pathway for disulfide bond formation in the endoplasmic reticulum.Mol Cell. 1999 Oct;4(4):469-77. doi: 10.1016/s1097-2765(00)80198-7. Mol Cell. 1999. PMID: 10549279
-
Oxidative protein folding: from thiol-disulfide exchange reactions to the redox poise of the endoplasmic reticulum.Free Radic Biol Med. 2015 Mar;80:171-82. doi: 10.1016/j.freeradbiomed.2014.07.037. Epub 2014 Aug 1. Free Radic Biol Med. 2015. PMID: 25091901 Free PMC article. Review.
-
Molecular mechanisms regulating oxidative activity of the Ero1 family in the endoplasmic reticulum.Antioxid Redox Signal. 2010 Oct;13(8):1177-87. doi: 10.1089/ars.2010.3230. Antioxid Redox Signal. 2010. PMID: 20486761 Review.
Cited by
-
A novel proteolytic event controls Hedgehog intracellular sorting and distribution to receptive fields.Biol Open. 2017 May 15;6(5):540-550. doi: 10.1242/bio.024083. Biol Open. 2017. PMID: 28298318 Free PMC article.
-
Redox-assisted protein folding systems in eukaryotic parasites.Antioxid Redox Signal. 2012 Aug 15;17(4):674-83. doi: 10.1089/ars.2011.4433. Epub 2012 Jan 10. Antioxid Redox Signal. 2012. PMID: 22122448 Free PMC article. Review.
-
Balanced Ero1 activation and inactivation establishes ER redox homeostasis.J Cell Biol. 2012 Mar 19;196(6):713-25. doi: 10.1083/jcb.201110090. Epub 2012 Mar 12. J Cell Biol. 2012. PMID: 22412017 Free PMC article.
-
Two phases of disulfide bond formation have differing requirements for oxygen.J Cell Biol. 2013 Nov 25;203(4):615-27. doi: 10.1083/jcb.201307185. Epub 2013 Nov 18. J Cell Biol. 2013. PMID: 24247433 Free PMC article.
-
Quantitative Analyses of the Yeast Oxidative Protein Folding Pathway In Vitro and In Vivo.Antioxid Redox Signal. 2019 Aug 1;31(4):261-274. doi: 10.1089/ars.2018.7615. Epub 2019 Apr 25. Antioxid Redox Signal. 2019. PMID: 30880408 Free PMC article.
References
-
- Frand A. R., Kaiser C. A. (1998) Mol. Cell 1, 161–170 - PubMed
-
- Pollard M. G., Travers K. J., Weissman J. S. (1998) Mol. Cell 1, 171–182 - PubMed
-
- Frand A. R., Kaiser C. A. (1999) Mol. Cell 4, 469–477 - PubMed
-
- Jessop C. E., Bulleid N. J. (2004) J. Biol. Chem. 279, 55341–55347 - PubMed
-
- Molteni S. N., Fassio A., Ciriolo M. R., Filomeni G., Pasqualetto E., Fagioli C., Sitia R. (2004) J. Biol. Chem. 279, 32667–32673 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

