Targeting of 5-aza-2'-deoxycytidine residues by chromatin-associated DNMT1 induces proteasomal degradation of the free enzyme
- PMID: 20348135
- PMCID: PMC2910061
- DOI: 10.1093/nar/gkq187
Targeting of 5-aza-2'-deoxycytidine residues by chromatin-associated DNMT1 induces proteasomal degradation of the free enzyme
Abstract
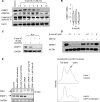
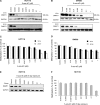
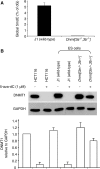
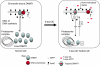
5-Aza-2'-deoxycytidine (5-aza-dC) is a nucleoside analogue with cytotoxic and DNA demethylating effects. Here we show that 5-aza-dC induces the proteasomal degradation of free (non-chromatin bound) DNMT1 through a mechanism which is dependent on DNA synthesis and the targeting of incorporated 5-aza-dC residues by DNMT1 itself. Thus, 5-aza-dC induces Dnmt1 degradation in wild-type mouse ES cells, but not in Dnmt [3a(-/-), 3b(-/-)] mouse ES cells which express Dnmt1 but lack DNA methylation (<0.7% of CpG methylated) and contain few hemi-methylated CpG sites, these being the preferred substrates for Dnmt1. We suggest that adducts formed between DNMT1 and 5-aza-dC molecules in DNA induce a ubiquitin-E3 ligase activity which preferentially targets free DNMT1 molecules for degradation by the proteasome. The proteasome inhibitor MG132 prevents DNMT1 degradation and reduces hypomethylation induced by 5-aza-dC.
Figures



References
-
- Taylor SM, Jones PA. Mechanism of action of eukaryotic DNA methyltransferase. Use of 5-azacytosine-containing DNA. J. Mol. Biol. 1982;162:679–692. - PubMed
-
- Wu JC, Santi DV. Kinetic and catalytic mechanism of HhaI methyltransferase. J. Biol. Chem. 1987;262:4778–4786. - PubMed
-
- Schermelleh L, Spada F, Easwaran HP, Zolghadr K, Margot JB, Cardoso MC, Leonhardt H. Trapped in action: direct visualization of DNA methyltransferase activity in living cells. Nat. Methods. 2005;2:751–756. - PubMed
-
- Ghoshal K, Datta J, Majumder S, Bai S, Kutay H, Motiwala T, Jacob ST. 5-Aza-deoxycytidine induces selective degradation of DNA methyltransferase 1 by a proteasomal pathway that requires the KEN box, bromo-adjacent homology domain, and nuclear localization signal. Mol. Cell. Biol. 2005;25:4727–4741. - PMC - PubMed
-
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

