Unusual evolution of a catalytic core element in CCA-adding enzymes
- PMID: 20348137
- PMCID: PMC2910056
- DOI: 10.1093/nar/gkq176
Unusual evolution of a catalytic core element in CCA-adding enzymes
Abstract
CCA-adding enzymes are polymerases existing in two distinct enzyme classes that both synthesize the C-C-A triplet at tRNA 3'-ends. Class II enzymes (found in bacteria and eukaryotes) carry a flexible loop in their catalytic core required for switching the specificity of the nucleotide binding pocket from CTP- to ATP-recognition. Despite this important function, the loop sequence varies strongly between individual class II CCA-adding enzymes. To investigate whether this loop operates as a discrete functional entity or whether it depends on the sequence context of the enzyme, we introduced reciprocal loop replacements in several enzymes. Surprisingly, many of these replacements are incompatible with enzymatic activity and inhibit ATP-incorporation. A phylogenetic analysis revealed the existence of conserved loop families. Loop replacements within families did not interfere with enzymatic activity, indicating that the loop function depends on a sequence context specific for individual enzyme families. Accordingly, modeling experiments suggest specific interactions of loop positions with important elements of the protein, forming a lever-like structure. Hence, although being part of the enzyme's catalytic core, the loop region follows an extraordinary evolutionary path, independent of other highly conserved catalytic core elements, but depending on specific sequence features in the context of the individual enzymes.
Figures

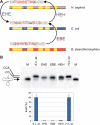
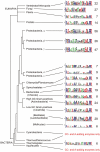
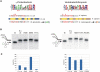
References
-
- Sprinzl M, Cramer F. The -C-C-A end of tRNA and its role in protein biosynthesis. Prog. Nucleic Acid Res. Mol. Biol. 1979;22:1–69. - PubMed
-
- Schimmel P, Yang XL. Two classes give lessons about CCA. Nat. Struct. Mol. Biol. 2004;11:807–808. - PubMed
-
- Weiner AM. tRNA maturation: RNA polymerization without a nucleic acid template. Curr. Biol. 2004;14:R883–R885. - PubMed
-
- Vörtler S, Mörl M. tRNA-nucleotidyltransferases: Highly unusual RNA polymerases with vital functions. FEBS lett. 2010;584:297–302. - PubMed

