The dual-targeted purple acid phosphatase isozyme AtPAP26 is essential for efficient acclimation of Arabidopsis to nutritional phosphate deprivation
- PMID: 20348213
- PMCID: PMC2899917
- DOI: 10.1104/pp.110.153270
The dual-targeted purple acid phosphatase isozyme AtPAP26 is essential for efficient acclimation of Arabidopsis to nutritional phosphate deprivation
Abstract
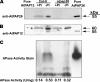
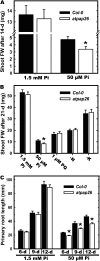
Induction of intracellular and secreted acid phosphatases (APases) is a widespread response of orthophosphate (Pi)-starved (-Pi) plants. APases catalyze Pi hydrolysis from a broad range of phosphomonoesters at an acidic pH. The largest class of nonspecific plant APases is comprised of the purple APases (PAPs). Although the biochemical properties, subcellular location, and expression of several plant PAPs have been described, their physiological functions have not been fully resolved. Recent biochemical studies indicated that AtPAP26, one of 29 PAPs encoded by the Arabidopsis (Arabidopsis thaliana) genome, is the predominant intracellular APase, as well as a major secreted APase isozyme up-regulated by -Pi Arabidopsis. An atpap26 T-DNA insertion mutant lacking AtPAP26 transcripts and 55-kD immunoreactive AtPAP26 polypeptides exhibited: (1) 9- and 5-fold lower shoot and root APase activity, respectively, which did not change in response to Pi starvation, (2) a 40% decrease in secreted APase activity during Pi deprivation, (3) 35% and 50% reductions in free and total Pi concentration, respectively, as well as 5-fold higher anthocyanin levels in shoots of soil-grown -Pi plants, and (4) impaired shoot and root development when subjected to Pi deficiency. By contrast, no deleterious influence of AtPAP26 loss of function occurred under Pi-replete conditions, or during nitrogen or potassium-limited growth, or oxidative stress. Transient expression of AtPAP26-mCherry in Arabidopsis suspension cells verified that AtPAP26 is targeted to the cell vacuole. Our results confirm that AtPAP26 is a principal contributor to Pi stress-inducible APase activity, and that it plays an important role in the Pi metabolism of -Pi Arabidopsis.
Figures




Similar articles
-
Biochemical and molecular characterization of AtPAP26, a vacuolar purple acid phosphatase up-regulated in phosphate-deprived Arabidopsis suspension cells and seedlings.Plant Physiol. 2006 Nov;142(3):1282-93. doi: 10.1104/pp.106.087171. Epub 2006 Sep 8. Plant Physiol. 2006. PMID: 16963519 Free PMC article.
-
The secreted purple acid phosphatase isozymes AtPAP12 and AtPAP26 play a pivotal role in extracellular phosphate-scavenging by Arabidopsis thaliana.J Exp Bot. 2012 Nov;63(18):6531-42. doi: 10.1093/jxb/ers309. Epub 2012 Nov 3. J Exp Bot. 2012. PMID: 23125358 Free PMC article.
-
Biochemical and molecular characterization of AtPAP12 and AtPAP26: the predominant purple acid phosphatase isozymes secreted by phosphate-starved Arabidopsis thaliana.Plant Cell Environ. 2010 Nov;33(11):1789-803. doi: 10.1111/j.1365-3040.2010.02184.x. Plant Cell Environ. 2010. PMID: 20545876
-
Functions and regulation of phosphate starvation-induced secreted acid phosphatases in higher plants.Plant Sci. 2018 Jun;271:108-116. doi: 10.1016/j.plantsci.2018.03.013. Epub 2018 Mar 14. Plant Sci. 2018. PMID: 29650148 Review.
-
The multifaceted nature of plant acid phosphatases: purification, biochemical features, and applications.J Enzyme Inhib Med Chem. 2023 Dec;38(1):2282379. doi: 10.1080/14756366.2023.2282379. Epub 2023 Nov 20. J Enzyme Inhib Med Chem. 2023. PMID: 37985663 Free PMC article. Review.
Cited by
-
The maize (Zea mays ssp. mays var. B73) genome encodes 33 members of the purple acid phosphatase family.Front Plant Sci. 2015 May 19;6:341. doi: 10.3389/fpls.2015.00341. eCollection 2015. Front Plant Sci. 2015. PMID: 26042133 Free PMC article.
-
Arabidopsis phosphatase under-producer mutants pup1 and pup3 contain mutations in the AtPAP10 and AtPAP26 genes.Plant Signal Behav. 2015;10(7):e1035851. doi: 10.1080/15592324.2015.1035851. Plant Signal Behav. 2015. PMID: 26251878 Free PMC article.
-
Characterization of purple acid phosphatases involved in extracellular dNTP utilization in Stylosanthes.J Exp Bot. 2016 Jul;67(14):4141-54. doi: 10.1093/jxb/erw190. Epub 2016 May 18. J Exp Bot. 2016. PMID: 27194738 Free PMC article.
-
Understanding the Adaptive Mechanisms of Plants to Enhance Phosphorus Use Efficiency on Podzolic Soils in Boreal Agroecosystems.Front Plant Sci. 2022 Mar 15;13:804058. doi: 10.3389/fpls.2022.804058. eCollection 2022. Front Plant Sci. 2022. PMID: 35371179 Free PMC article. Review.
-
Plant ecological genomics at the limits of life in the Atacama Desert.Proc Natl Acad Sci U S A. 2021 Nov 16;118(46):e2101177118. doi: 10.1073/pnas.2101177118. Proc Natl Acad Sci U S A. 2021. PMID: 34725254 Free PMC article.
References
-
- Alonso JM, Stepanova A, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 - PubMed
-
- Bozzo GG, Dunn EL, Plaxton WC. (2006) Differential synthesis of phosphate-starvation inducible purple acid phosphatase isozymes in tomato (Lycopersicon esculentum) suspension cells and seedlings. Plant Cell Environ 29: 303–313 - PubMed
-
- Bozzo GG, Plaxton WC. (2008) The role of intracellular and secreted purple acid phosphatases in tomato phosphate nutrition. Preddy V, Watson R, , Tomatoes and Tomato Products. Science Publishers, Enfield, NH, pp 216–233
-
- Bozzo GG, Raghothama KG, Plaxton WC. (2002) Purification and characterization of two secreted purple acid phosphatase isozymes from phosphate-starved tomato (Lycopersicon esculentum) cell cultures. Eur J Biochem 269: 6278–6286 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

