Biochemical characterization of a novel indole prenyltransferase from Streptomyces sp. SN-593
- PMID: 20348259
- PMCID: PMC2876496
- DOI: 10.1128/JB.01557-09
Biochemical characterization of a novel indole prenyltransferase from Streptomyces sp. SN-593
Abstract
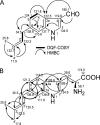
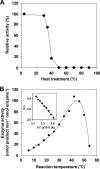
Genome sequencing of Streptomyces species has highlighted numerous potential genes of secondary metabolite biosynthesis. The mining of cryptic genes is important for exploring chemical diversity. Here we report the metabolite-guided genome mining and functional characterization of a cryptic gene by biochemical studies. Based on systematic purification of metabolites from Streptomyces sp. SN-593, we isolated a novel compound, 6-dimethylallylindole (DMAI)-3-carbaldehyde. Although many 6-DMAI compounds have been isolated from a variety of organisms, an enzyme catalyzing the transfer of a dimethylallyl group to the C-6 indole ring has not been reported so far. A homology search using known prenyltransferase sequences against the draft sequence of the Streptomyces sp. SN-593 genome revealed the iptA gene. The IptA protein showed 27% amino acid identity to cyanobacterial LtxC, which catalyzes the transfer of a geranyl group to (-)-indolactam V. A BLAST search against IptA revealed much-more-similar homologs at the amino acid level than LtxC, namely, SAML0654 (60%) from Streptomyces ambofaciens ATCC 23877 and SCO7467 (58%) from S. coelicolor A3(2). Phylogenetic analysis showed that IptA was distinct from bacterial aromatic prenyltransferases and fungal indole prenyltransferases. Detailed kinetic analyses of IptA showed the highest catalytic efficiency (6.13 min(-1) microM(-1)) for L-Trp in the presence of dimethylallyl pyrophosphate (DMAPP), suggesting that the enzyme is a 6-dimethylallyl-L-Trp synthase (6-DMATS). Substrate specificity analyses of IptA revealed promiscuity for indole derivatives, and its reaction products were identified as novel 6-DMAI compounds. Moreover, DeltaiptA mutants abolished the production of 6-DMAI-3-carbaldehyde as well as 6-dimethylallyl-L-Trp, suggesting that the iptA gene is involved in the production of 6-DMAI-3-carbaldehyde.
Figures






Similar articles
-
Crystal structures of a 6-dimethylallyltryptophan synthase, IptA: Insights into substrate tolerance and enhancement of prenyltransferase activity.Biochem Biophys Res Commun. 2022 Feb 19;593:144-150. doi: 10.1016/j.bbrc.2022.01.018. Epub 2022 Jan 12. Biochem Biophys Res Commun. 2022. PMID: 35074664
-
Isolation, structural elucidation and biosynthesis of 3-hydroxy-6-dimethylallylindolin-2-one, a novel prenylated indole derivative from Actinoplanes missouriensis.J Antibiot (Tokyo). 2014 Mar;67(3):231-6. doi: 10.1038/ja.2013.116. Epub 2013 Nov 13. J Antibiot (Tokyo). 2014. PMID: 24220111
-
Biochemical investigations of two 6-DMATS enzymes from Streptomyces reveal new features of L-tryptophan prenyltransferases.Chembiochem. 2014 May 5;15(7):1030-9. doi: 10.1002/cbic.201400046. Epub 2014 Apr 1. Chembiochem. 2014. PMID: 24692239
-
Indole prenyltransferases from fungi: a new enzyme group with high potential for the production of prenylated indole derivatives.Curr Med Chem. 2009;16(2):218-31. doi: 10.2174/092986709787002772. Curr Med Chem. 2009. PMID: 19149573 Review.
-
Biological matching of chemical reactivity: pairing indole nucleophilicity with electrophilic isoprenoids.ACS Chem Biol. 2014 Dec 19;9(12):2718-28. doi: 10.1021/cb500695k. Epub 2014 Oct 29. ACS Chem Biol. 2014. PMID: 25303280 Review.
Cited by
-
Functional characterization of the promiscuous prenyltransferase responsible for furaquinocin biosynthesis: identification of a physiological polyketide substrate and its prenylated reaction products.J Biol Chem. 2010 Dec 17;285(51):39663-71. doi: 10.1074/jbc.M110.153957. Epub 2010 Oct 11. J Biol Chem. 2010. PMID: 20937800 Free PMC article.
-
Biochemical characterization of indole prenyltransferases: filling the last gap of prenylation positions by a 5-dimethylallyltryptophan synthase from Aspergillus clavatus.J Biol Chem. 2012 Jan 6;287(2):1371-80. doi: 10.1074/jbc.M111.317982. Epub 2011 Nov 28. J Biol Chem. 2012. PMID: 22123822 Free PMC article.
-
The biosynthetic genes for prenylated phenazines are located at two different chromosomal loci of Streptomyces cinnamonensis DSM 1042.Microb Biotechnol. 2011 Mar;4(2):252-62. doi: 10.1111/j.1751-7915.2010.00234.x. Epub 2010 Dec 8. Microb Biotechnol. 2011. PMID: 21342470 Free PMC article.
-
The tRNA-dependent biosynthesis of modified cyclic dipeptides.Int J Mol Sci. 2014 Aug 21;15(8):14610-31. doi: 10.3390/ijms150814610. Int J Mol Sci. 2014. PMID: 25196600 Free PMC article. Review.
-
Multisite prenylation of 4-substituted tryptophans by dimethylallyltryptophan synthase.J Am Chem Soc. 2013 Feb 6;135(5):1895-902. doi: 10.1021/ja310734n. Epub 2013 Jan 28. J Am Chem Soc. 2013. PMID: 23301871 Free PMC article.
References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Archenbach, H., C. Renner, and R. Waibel. 1995. The hexalobines, diprenylated indoles from Hexalobus crispiflorus and Hexalobus monopetalus. Liebigs Ann. 1995:1327-1337.
-
- Archenbach, H., and B. Raffelsberger. 1979. 3,6-Bis(γ,γ-dimethylallyl)-indole from Uvaria elliotiana. Tetrahedron Lett. 28:2571-2574.
-
- Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. - PubMed
-
- Bergmann, S., J. Schumann, K. Scherlach, C. Lange, A. A. Brakhage, and C. Hertweck. 2007. Genomics-driven discovery of PKS-NRPS hybrid metabolites from Aspergillus nidulans. Nat. Chem. Biol. 3:213-217. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

