Negative regulation of expression of the nitrate assimilation nirA operon in the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120
- PMID: 20348260
- PMCID: PMC2876489
- DOI: 10.1128/JB.01668-09
Negative regulation of expression of the nitrate assimilation nirA operon in the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120
Abstract
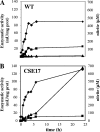
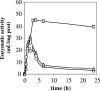
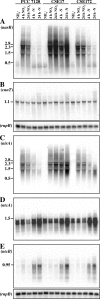
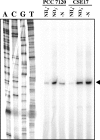
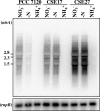
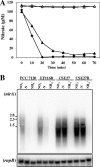
In the filamentous, heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120, expression of the nitrate assimilation nirA operon takes place in the absence of ammonium and the presence of nitrate or nitrite. Several positive-action proteins that are required for expression of the nirA operon have been identified. Whereas NtcA and NtcB exert their action by direct binding to the nirA operon promoter, CnaT acts by an as yet unknown mechanism. In the genome of this cyanobacterium, open reading frame (ORF) all0605 (the nirB gene) is found between the nirA (encoding nitrite reductase) and ntcB genes. A nirB mutant was able to grow at the expense of nitrate as a nitrogen source and showed abnormally high levels of nirA operon mRNA both in the presence and in the absence of nitrate. This mutant showed increased nitrate reductase activity but decreased nitrite reductase activity, an imbalance that resulted in excretion of nitrite, which accumulated in the extracellular medium, when the nirB mutant was grown in the presence of nitrate. A nirA in-frame deletion mutant also showed a phenotype of increased expression of the nirA operon in the absence of ammonium, independent of the presence of nitrate in the medium. Both NirB and NirA are therefore needed to keep low levels of expression of the nirA operon in the absence of an inducer. Because NirB is also needed to attain high levels of nitrite reductase activity, NirA appears to be a negative element in the nitrate regulation of expression of the nirA operon in Anabaena sp. strain PCC 7120.
Figures

References
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 2009. Current protocols in molecular biology. Greene Publishing and Wiley-Interscience, New York, NY.
-
- Black, T. A., Y. Cai, and C. P. Wolk. 1993. Spatial expression and autoregulation of hetR, a gene involved in the control of heterocyst development in Anabaena. Mol. Microbiol. 9:77-84. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

