Cloning of separate meilingmycin biosynthesis gene clusters by use of acyltransferase-ketoreductase didomain PCR amplification
- PMID: 20348291
- PMCID: PMC2869129
- DOI: 10.1128/AEM.02262-09
Cloning of separate meilingmycin biosynthesis gene clusters by use of acyltransferase-ketoreductase didomain PCR amplification
Abstract
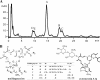
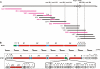
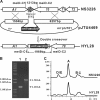
Five meilingmycins, A to E, with A as the major component, were isolated from Streptomyces nanchangensis NS3226. Through nuclear magnetic resonance (NMR) characterization, meilingmycins A to E proved to be identical to reported milbemycins alpha11, alpha13, alpha14, beta1, and beta9, respectively. Sequencing of a previously cloned 103-kb region identified three modular type I polyketide synthase genes putatively encoding the last 11 elongation steps, three modification proteins, and one transcriptional regulatory protein for meilingmycin biosynthesis. However, the expected loading module and the first two elongation modules were missing. In meilingmycin, the presence of a methyl group at C-24 and a hydroxyl group at C-25 suggests that the elongation module 1 contains a methylmalonyl-coenzyme A (CoA)-specific acyltransferase (ATp) domain and a ketoreductase (KR) domain. Based on the conserved motifs of the ATp and KR domains, a pair of primers was designed for PCR amplification, and a 1.40-kb expected fragment was amplified, whose sequence shows significant homology with the elongation module 1 of the aveA1-encoded enzyme AVES1. A polyketide synthase (PKS) gene encoding one loading and two elongation modules, with a downstream C-5-O-methyltransferase gene, meiD, was subsequently localized 55 kb apart from the previously sequenced region, and its deletion abolishes meilingmycin production. A series of deletions within the 55-kb intercluster region rules out its involvement in meilingmycin biosynthesis. Furthermore, gene deletion of meiD eliminates meilingmycins D and E, with methyls at C-5. Our work provides a more specific strategy for the cloning of modular type I PKS gene clusters. The cloning of the meilingmycin gene clusters paves the way for its pathway engineering.
Figures



Similar articles
-
Identification of a gene cluster encoding meilingmycin biosynthesis among multiple polyketide synthase contigs isolated from Streptomyces nanchangensis NS3226.Arch Microbiol. 2003 Aug;180(2):101-7. doi: 10.1007/s00203-003-0564-1. Epub 2003 Jun 14. Arch Microbiol. 2003. PMID: 12811466
-
The biosynthetic gene cluster for the antitumor drug bleomycin from Streptomyces verticillus ATCC15003 supporting functional interactions between nonribosomal peptide synthetases and a polyketide synthase.Chem Biol. 2000 Aug;7(8):623-42. doi: 10.1016/s1074-5521(00)00011-9. Chem Biol. 2000. PMID: 11048953
-
Characterization of the enzymatic domains in the modular polyketide synthase involved in rifamycin B biosynthesis by Amycolatopsis mediterranei.Gene. 1998 Aug 31;216(2):255-65. doi: 10.1016/s0378-1119(98)00338-2. Gene. 1998. PMID: 9729415
-
Characterization of biosynthetic gene cluster for the production of virginiamycin M, a streptogramin type A antibiotic, in Streptomyces virginiae.Gene. 2007 May 15;393(1-2):31-42. doi: 10.1016/j.gene.2006.12.035. Epub 2007 Jan 20. Gene. 2007. PMID: 17350183
-
Cloning, sequencing and analysis of the enterocin biosynthesis gene cluster from the marine isolate 'Streptomyces maritimus': evidence for the derailment of an aromatic polyketide synthase.Chem Biol. 2000 Dec;7(12):943-55. doi: 10.1016/s1074-5521(00)00044-2. Chem Biol. 2000. PMID: 11137817
Cited by
-
MilR3, a unique SARP family pleiotropic regulator in Streptomyces bingchenggensis.Arch Microbiol. 2022 Sep 19;204(10):631. doi: 10.1007/s00203-022-03240-x. Arch Microbiol. 2022. PMID: 36121479
-
Recent advances in the research of milbemycin biosynthesis and regulation as well as strategies for strain improvement.Arch Microbiol. 2021 Dec;203(10):5849-5857. doi: 10.1007/s00203-021-02575-1. Epub 2021 Sep 22. Arch Microbiol. 2021. PMID: 34550409 Review.
-
Engineering of primary metabolic pathways for titer improvement of milbemycins in Streptomyces bingchenggensis.Appl Microbiol Biotechnol. 2021 Mar;105(5):1875-1887. doi: 10.1007/s00253-021-11164-7. Epub 2021 Feb 10. Appl Microbiol Biotechnol. 2021. PMID: 33564920
-
Overproduction of endusamycin in Streptomyces endus subsp. aureus.Synth Syst Biotechnol. 2025 Feb 14;10(2):523-531. doi: 10.1016/j.synbio.2025.02.004. eCollection 2025 Jun. Synth Syst Biotechnol. 2025. PMID: 40061181 Free PMC article.
-
Cytochromes P450 for natural product biosynthesis in Streptomyces: sequence, structure, and function.Nat Prod Rep. 2017 Aug 30;34(9):1141-1172. doi: 10.1039/c7np00034k. Nat Prod Rep. 2017. PMID: 28758170 Free PMC article. Review.
References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Burg, R. W., B. M. Miller, E. E. Baker, J. Birnbaum, S. A. Currie, R. Hartman, Y. L. Kong, R. L. Monaghan, G. Olson, I. Putter, J. B. Tunac, H. Wallick, E. O. Stapley, R. Oiwa, and S. Omura. 1979. Avermectins, new family of potent anthelmintic agents: producing organism and fermentation. Antimicrob. Agents Chemother. 15:361-367. - PMC - PubMed
-
- Caffrey, P. 2003. Conserved amino acid residues correlating with ketoreductase stereospecificity in modular polyketide synthases. Chembiochem 4:654-657. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

