Characterization of the serpin-encoding gene of Bifidobacterium breve 210B
- PMID: 20348296
- PMCID: PMC2869134
- DOI: 10.1128/AEM.02938-09
Characterization of the serpin-encoding gene of Bifidobacterium breve 210B
Abstract
Members of the serpin (serine protease inhibitor) superfamily have been identified in higher multicellular eukaryotes, as well as in bacteria, although examination of available genome sequences has indicated that homologs of the bacterial serpin-encoding gene (ser) are not widely distributed. In members of the genus Bifidobacterium this gene appears to be present in at least 5, and perhaps up to 9, of the 30 species tested. Moreover, phylogenetic analysis using available bacterial and eukaryotic serpin sequences revealed that bifidobacteria produce serpins that form a separate clade. We characterized the ser(210B) locus of Bifidobacterium breve 210B, which encompasses a number of genes whose deduced protein products display significant similarity to proteins encoded by corresponding loci found in several other bifidobacteria. Northern hybridization, primer extension, microarray, reverse transcription-PCR (RT-PCR), and quantitative real-time PCR (qRT-PCR) analyses revealed that a 3.5-kb polycistronic mRNA encompassing the ser(210B) operon with a single transcriptional start site is strongly induced following treatment of B. breve 210B cultures with some proteases. Interestingly, transcription of other bifidobacterial ser homologs appears to be triggered by different proteases.
Figures





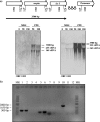

Similar articles
-
Gene structure and transcriptional organization of the dnaK operon of Bifidobacterium breve UCC 2003 and application of the operon in bifidobacterial tracing.Appl Environ Microbiol. 2005 Jan;71(1):487-500. doi: 10.1128/AEM.71.1.487-500.2005. Appl Environ Microbiol. 2005. PMID: 15640225 Free PMC article.
-
Molecular characterization of hsp20, encoding a small heat shock protein of bifidobacterium breve UCC2003.Appl Environ Microbiol. 2007 Jul;73(14):4695-703. doi: 10.1128/AEM.02496-06. Epub 2007 May 18. Appl Environ Microbiol. 2007. PMID: 17513584 Free PMC article.
-
A two-component regulatory system controls autoregulated serpin expression in Bifidobacterium breve UCC2003.Appl Environ Microbiol. 2012 Oct;78(19):7032-41. doi: 10.1128/AEM.01776-12. Epub 2012 Jul 27. Appl Environ Microbiol. 2012. PMID: 22843530 Free PMC article.
-
Genus- and species-specific PCR primers for the detection and identification of bifidobacteria.Curr Issues Intest Microbiol. 2003 Sep;4(2):61-9. Curr Issues Intest Microbiol. 2003. PMID: 14503690 Review.
-
Genomics as a means to understand bacterial phylogeny and ecological adaptation: the case of bifidobacteria.Antonie Van Leeuwenhoek. 2007 May;91(4):351-72. doi: 10.1007/s10482-006-9122-6. Epub 2006 Oct 28. Antonie Van Leeuwenhoek. 2007. PMID: 17072531 Review.
Cited by
-
Bifidobacterial Dialogue With Its Human Host and Consequent Modulation of the Immune System.Front Immunol. 2019 Oct 1;10:2348. doi: 10.3389/fimmu.2019.02348. eCollection 2019. Front Immunol. 2019. PMID: 31632412 Free PMC article. Review.
-
Genomic characterization and transcriptional studies of the starch-utilizing strain Bifidobacterium adolescentis 22L.Appl Environ Microbiol. 2014 Oct;80(19):6080-90. doi: 10.1128/AEM.01993-14. Epub 2014 Jul 25. Appl Environ Microbiol. 2014. PMID: 25063659 Free PMC article.
-
The First Microbial Colonizers of the Human Gut: Composition, Activities, and Health Implications of the Infant Gut Microbiota.Microbiol Mol Biol Rev. 2017 Nov 8;81(4):e00036-17. doi: 10.1128/MMBR.00036-17. Print 2017 Dec. Microbiol Mol Biol Rev. 2017. PMID: 29118049 Free PMC article. Review.
-
Genomic analysis of three Bifidobacterium species isolated from the calf gastrointestinal tract.Sci Rep. 2016 Jul 29;6:30768. doi: 10.1038/srep30768. Sci Rep. 2016. PMID: 27468806 Free PMC article.
-
Gut Serpinome: Emerging Evidence in IBD.Int J Mol Sci. 2021 Jun 4;22(11):6088. doi: 10.3390/ijms22116088. Int J Mol Sci. 2021. PMID: 34200095 Free PMC article. Review.
References
-
- Barrangou, R., E. P. Briczinski, L. L. Traeger, J. R. Loquasto, M. Richards, P. Horvath, A. C. Coute-Monvoisin, G. Leyer, S. Rendulic, J. L. Steele, J. R. Broadbent, T. Oberg, E. G. Dudley, S. Schuster, D. A. Romero, and R. F. Roberts. 2009. Comparison of the complete genome sequences of Bifidobacterium animalis subsp. lactis DSM 10140 and Bl-04. J. Bacteriol. 191:4144-4151. - PMC - PubMed
-
- Benjamini, Y., and Y. Hochberg. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. 57:289-300.
-
- Bolstad, B. M., R. A. Irizarry, M. Astrand, and T. P. Speed. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19:185-193. - PubMed
-
- Burg, N. D., and M. H. Pillinger. 2001. The neutrophil: function and regulation in innate and humoral immunity. Clin. Immunol. 99:7-17. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

