Constitutive secretion in Tetrahymena thermophila
- PMID: 20348385
- PMCID: PMC2863955
- DOI: 10.1128/EC.00024-10
Constitutive secretion in Tetrahymena thermophila
Abstract
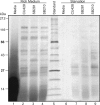
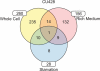
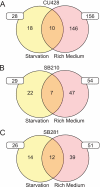
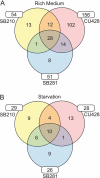
The growth, survival, and life cycle progression of the freshwater ciliated protozoan Tetrahymena thermophila are responsive to protein signals thought to be released by constitutive secretion. In addition to providing insights about ciliate communication, studies of constitutive secretion are of interest for evaluating the utility of T. thermophila as a platform for the expression of secreted protein therapeutics. For these reasons, we undertook an unbiased investigation of T. thermophila secreted proteins using wild-type and secretion mutant strains. Extensive tandem mass spectrometry analyses of secretome samples were performed. We identified a total of 207 secretome proteins, most of which were not detected in a set of abundant whole-cell protein identifications. Numerous proteases and other hydrolases were secreted from cells grown in rich medium but not cells transferred to a nutrient starvation condition. On the other hand, we detected the starvation-enhanced secretion of a small number of cytosolic proteins, suggestive of an exosome-like pathway in T. thermophila. Subsets of proteins from the T. thermophila regulated secretion pathway were detected with differential representation across strains and culture conditions. Finally, many secretome proteins had a predicted N-terminal signal sequence but no other annotated characteristic or functional classification. Our work provides the first comprehensive analysis of secreted proteins in T. thermophila and establishes the groundwork for future studies of constitutive protein secretion biology and biotechnology in ciliates.
Figures
Similar articles
-
Functional proteomics protocol for the identification of interaction partners in Tetrahymena thermophila.STAR Protoc. 2021 Mar 4;2(1):100362. doi: 10.1016/j.xpro.2021.100362. eCollection 2021 Mar 19. STAR Protoc. 2021. PMID: 33786459 Free PMC article.
-
Expression, secretion and surface display of a human alkaline phosphatase by the ciliate Tetrahymena thermophila.BMC Biotechnol. 2011 Jan 31;11:11. doi: 10.1186/1472-6750-11-11. BMC Biotechnol. 2011. PMID: 21281462 Free PMC article.
-
Nucleus-specific linker histones Hho1 and Mlh1 form distinct protein interactions during growth, starvation and development in Tetrahymena thermophila.Sci Rep. 2020 Jan 13;10(1):168. doi: 10.1038/s41598-019-56867-0. Sci Rep. 2020. PMID: 31932604 Free PMC article.
-
Regulated protein secretion in Tetrahymena thermophila.Methods Cell Biol. 2000;62:347-62. doi: 10.1016/s0091-679x(08)61542-3. Methods Cell Biol. 2000. PMID: 10503203 Review. No abstract available.
-
Bromodomain-containing proteins in the unicellular eukaryote Tetrahymena thermophila.Zool Res. 2025 May 18;46(3):538-550. doi: 10.24272/j.issn.2095-8137.2025.011. Zool Res. 2025. PMID: 40259734 Review.
Cited by
-
Inferring gene-pathway associations from consolidated transcriptome datasets: an interactive gene network explorer for Tetrahymena thermophila.NAR Genom Bioinform. 2025 May 27;7(2):lqaf067. doi: 10.1093/nargab/lqaf067. eCollection 2025 Jun. NAR Genom Bioinform. 2025. PMID: 40432793 Free PMC article.
-
Membrane dynamics at the nuclear exchange junction during early mating (one to four hours) in the ciliate Tetrahymena thermophila.Eukaryot Cell. 2015 Feb;14(2):116-27. doi: 10.1128/EC.00164-14. Epub 2014 Aug 8. Eukaryot Cell. 2015. PMID: 25107923 Free PMC article.
-
The secretory pathway in Tetrahymena is organized for efficient constitutive secretion at ciliary pockets.iScience. 2024 Oct 9;27(11):111123. doi: 10.1016/j.isci.2024.111123. eCollection 2024 Nov 15. iScience. 2024. PMID: 39498308 Free PMC article.
-
Comprehensive analysis reveals dynamic and evolutionary plasticity of Rab GTPases and membrane traffic in Tetrahymena thermophila.PLoS Genet. 2010 Oct 14;6(10):e1001155. doi: 10.1371/journal.pgen.1001155. PLoS Genet. 2010. PMID: 20976245 Free PMC article.
-
Conservation and innovation in Tetrahymena membrane traffic: proteins, lipids, and compartments.Methods Cell Biol. 2012;109:141-75. doi: 10.1016/B978-0-12-385967-9.00006-2. Methods Cell Biol. 2012. PMID: 22444145 Free PMC article. Review.
References
-
- Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T., Harris M. A., Hill D. P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J. C., Richardson J. E., Ringwald M., Rubin G. M., Sherlock G. 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25:25–29 - PMC - PubMed
-
- Bouws H., Wattenberg A., Zorn H. 2008. Fungal secretomes—nature's toolbox for white biotechnology. Appl. Microbiol. Biotechnol. 80:381–388 - PubMed
-
- Bowman G. R., Smith D. G., Michael Siu K. W., Pearlman R. E., Turkewitz A. P. 2005. Genomic and proteomic evidence for a second family of dense core granule cargo proteins in Tetrahymena thermophila. J. Eukaryot. Microbiol. 52:291–297 - PubMed
-
- Brambilla F., Resta D., Isak I., Zanotti M., Arnoldi A. 2009. A label-free internal standard method for the differential analysis of bioactive lupin proteins using nano HPLC-chip coupled with ion trap mass spectrometry. Proteomics 9:272–286 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

