Alkylresorcinol synthases expressed in Sorghum bicolor root hairs play an essential role in the biosynthesis of the allelopathic benzoquinone sorgoleone
- PMID: 20348430
- PMCID: PMC2861460
- DOI: 10.1105/tpc.109.072397
Alkylresorcinol synthases expressed in Sorghum bicolor root hairs play an essential role in the biosynthesis of the allelopathic benzoquinone sorgoleone
Abstract
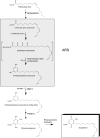
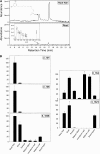
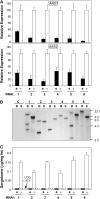
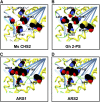
Sorghum bicolor is considered to be an allelopathic crop species, producing phytotoxins such as the lipid benzoquinone sorgoleone, which likely accounts for many of the allelopathic properties of Sorghum spp. Current evidence suggests that sorgoleone biosynthesis occurs exclusively in root hair cells and involves the production of an alkylresorcinolic intermediate (5-[(Z,Z)-8',11',14'-pentadecatrienyl]resorcinol) derived from an unusual 16:3Delta(9,12,15) fatty acyl-CoA starter unit. This led to the suggestion of the involvement of one or more alkylresorcinol synthases (ARSs), type III polyketide synthases (PKSs) that produce 5-alkylresorcinols using medium to long-chain fatty acyl-CoA starter units via iterative condensations with malonyl-CoA. In an effort to characterize the enzymes responsible for the biosynthesis of the pentadecyl resorcinol intermediate, a previously described expressed sequence tag database prepared from isolated S. bicolor (genotype BTx623) root hairs was first mined for all PKS-like sequences. Quantitative real-time RT-PCR analyses revealed that three of these sequences were preferentially expressed in root hairs, two of which (designated ARS1 and ARS2) were found to encode ARS enzymes capable of accepting a variety of fatty acyl-CoA starter units in recombinant enzyme studies. Furthermore, RNA interference experiments directed against ARS1 and ARS2 resulted in the generation of multiple independent transformant events exhibiting dramatically reduced sorgoleone levels. Thus, both ARS1 and ARS2 are likely to participate in the biosynthesis of sorgoleone in planta. The sequences of ARS1 and ARS2 were also used to identify several rice (Oryza sativa) genes encoding ARSs, which are likely involved in the production of defense-related alkylresorcinols.
Figures





References
-
- Abe I., Morita H., Oguro S., Noma H., Wanibuchi K., Kawahara N., Goda Y., Noguchi H., Kohno T. (2007). Structure-based engineering of a plant type III polyketide synthase: Formation of an unnatural nonaketide naphthopyrone. J. Am. Chem. Soc. 129: 5976–5980 - PubMed
-
- Abe I., Oguro S., Utsumi Y., Sano Y., Noguchi H. (2005a). Engineered biosynthesis of plant polyketides: Chain length control in an octaketide-producing plant type III polyketide synthase. J. Am. Chem. Soc. 127: 12709–12716 - PubMed
-
- Abe I., Utsumi Y., Oguro S., Morita H., Sano Y., Noguchi H. (2005b). A plant type III polyketide synthase that produces pentaketide chromone. J. Am. Chem. Soc. 127: 1362–1363 - PubMed
-
- Abe I., Utsumi Y., Oguro S., Noguchi H. (2004). The first plant type III polyketide synthase that catalyzes formation of aromatic heptaketide. FEBS Lett. 562: 171–176 - PubMed
-
- Abe I., Watanabe T., Lou W., Noguchi H. (2006). Active site residues governing substrate selectivity and polyketide chain length in aloesone synthase. FEBS J. 273: 208–218 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

