Innate immune responses activated in Arabidopsis roots by microbe-associated molecular patterns
- PMID: 20348432
- PMCID: PMC2861455
- DOI: 10.1105/tpc.109.069658
Innate immune responses activated in Arabidopsis roots by microbe-associated molecular patterns
Abstract
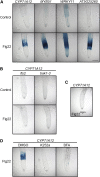
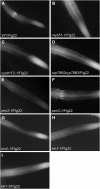
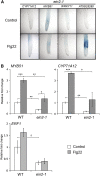
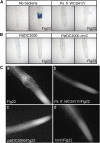
Despite the fact that roots are the organs most subject to microbial interactions, very little is known about the response of roots to microbe-associated molecular patterns (MAMPs). By monitoring transcriptional activation of beta-glucuronidase reporters and MAMP-elicited callose deposition, we show that three MAMPs, the flagellar peptide Flg22, peptidoglycan, and chitin, trigger a strong tissue-specific response in Arabidopsis thaliana roots, either at the elongation zone for Flg22 and peptidoglycan or in the mature parts of the roots for chitin. Ethylene signaling, the 4-methoxy-indole-3-ylmethylglucosinolate biosynthetic pathway, and the PEN2 myrosinase, but not salicylic acid or jasmonic acid signaling, play major roles in this MAMP response. We also show that Flg22 induces the cytochrome P450 CYP71A12-dependent exudation of the phytoalexin camalexin by Arabidopsis roots. The phytotoxin coronatine, an Ile-jasmonic acid mimic produced by Pseudomonas syringae pathovars, suppresses MAMP-activated responses in the roots. This suppression requires the E3 ubiquitin ligase COI1 as well as the transcription factor JIN1/MYC2 but does not rely on salicylic acid-jasmonic acid antagonism. These experiments demonstrate the presence of highly orchestrated and tissue-specific MAMP responses in roots and potential pathogen-encoded mechanisms to block these MAMP-elicited signaling pathways.
Figures







Similar articles
-
Microbe-associated molecular patterns-triggered root responses mediate beneficial rhizobacterial recruitment in Arabidopsis.Plant Physiol. 2012 Nov;160(3):1642-61. doi: 10.1104/pp.112.200386. Epub 2012 Sep 12. Plant Physiol. 2012. PMID: 22972705 Free PMC article.
-
The coronatine toxin of Pseudomonas syringae is a multifunctional suppressor of Arabidopsis defense.Plant Cell. 2012 Nov;24(11):4763-74. doi: 10.1105/tpc.112.105312. Epub 2012 Nov 30. Plant Cell. 2012. PMID: 23204405 Free PMC article.
-
The activated SA and JA signaling pathways have an influence on flg22-triggered oxidative burst and callose deposition.PLoS One. 2014 Feb 25;9(2):e88951. doi: 10.1371/journal.pone.0088951. eCollection 2014. PLoS One. 2014. PMID: 24586453 Free PMC article.
-
Responses of Arabidopsis thaliana to challenge by Pseudomonas syringae.Mol Cells. 2008 May 31;25(3):323-31. Epub 2008 May 16. Mol Cells. 2008. PMID: 18483469 Review.
-
Chitin and chitin-related compounds in plant-fungal interactions.Mycology. 2018 May 15;9(3):189-201. doi: 10.1080/21501203.2018.1473299. eCollection 2018. Mycology. 2018. PMID: 30181925 Free PMC article. Review.
Cited by
-
Transcriptome analysis unravels the biocontrol mechanism of Serratia plymuthica A30 against potato soft rot caused by Dickeya solani.PLoS One. 2024 Sep 6;19(9):e0308744. doi: 10.1371/journal.pone.0308744. eCollection 2024. PLoS One. 2024. PMID: 39240997 Free PMC article.
-
Lysin motif-containing proteins LYP4 and LYP6 play dual roles in peptidoglycan and chitin perception in rice innate immunity.Plant Cell. 2012 Aug;24(8):3406-19. doi: 10.1105/tpc.112.102475. Epub 2012 Aug 7. Plant Cell. 2012. PMID: 22872757 Free PMC article.
-
Deciphering the role of rhizosphere microbiota in modulating disease resistance in cabbage varieties.Microbiome. 2024 Aug 30;12(1):160. doi: 10.1186/s40168-024-01883-0. Microbiome. 2024. PMID: 39215347 Free PMC article.
-
Arabidopsis NATA1 Acetylates Putrescine and Decreases Defense-Related Hydrogen Peroxide Accumulation.Plant Physiol. 2016 Jun;171(2):1443-55. doi: 10.1104/pp.16.00446. Epub 2016 Apr 25. Plant Physiol. 2016. PMID: 27208290 Free PMC article.
-
Seaweed Extract (Stella Maris®) Activates Innate Immune Responses in Arabidopsis thaliana and Protects Host against Bacterial Pathogens.Mar Drugs. 2018 Jun 28;16(7):221. doi: 10.3390/md16070221. Mar Drugs. 2018. PMID: 29958402 Free PMC article.
References
-
- Aist J.R., Bushnell W.R. (1991). Invasion of plants by powdery mildew fungi, and cellular mechanisms of resistance. In The Fungal Spore and Disease Initiation in Plants and Animals, Cole G.T., Mock H.C., (New York: Plenum; ), pp. 321–345
-
- Andreote F.D., de Araujo W.L., de Azevedo J.L., van Elsas J.D., da Rocha U.N., van Overbeek L.S. (2009). Endophytic colonization of potato (Solanum tuberosum L.) by a novel competent bacterial endophyte, Pseudomonas putida strain P9, and its effect on associated bacterial communities. Appl. Environ. Microbiol. 75: 3396–3406 - PMC - PubMed
-
- Attard A., Gourgues M., Galiana E., Panabieres F., Ponchet M., Keller H. (2008). Strategies of attack and defense in plant-oomycete interactions, accentuated for Phytophthora parasitica Dastur (syn. P. Nicotianae Breda de Haan). J. Plant Physiol. 165: 83–94 - PubMed
-
- Badri D.V., Vivanco J.M. (2009). Regulation and function of root exudates. Plant Cell Environ. 32: 666–681 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

