Selection of RNA aptamers imported into yeast and human mitochondria
- PMID: 20348443
- PMCID: PMC2856887
- DOI: 10.1261/rna.1914110
Selection of RNA aptamers imported into yeast and human mitochondria
Abstract
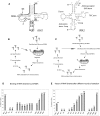
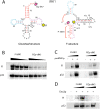
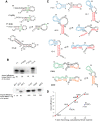
In the yeast Saccharomyces cerevisiae, nuclear DNA-encoded is partially imported into mitochondria. We previously found that the synthetic transcripts of yeast tRNA(Lys) and a number of their mutant versions could be specifically internalized by isolated yeast and human mitochondria. The mitochondrial targeting of tRNA(Lys) in yeast was shown to depend on the cytosolic precursor of mitochondrial lysyl-tRNA synthetase and the glycolytic enzyme enolase. Here we applied the approach of in vitro selection (SELEX) to broaden the spectrum of importable tRNA-derived molecules. We found that RNAs selected for their import into isolated yeast mitochondria have lost the potential to acquire a classical tRNA-shape. Analysis of conformational rearrangements in the importable RNAs by in-gel fluorescence resonance energy transfer (FRET) approach permitted us to suggest that protein factor binding and subsequent import require formation of an alternative structure, different from a classic L-form tRNA model. We show that in the complex with targeting protein factor, enolase 2, tRK1 adopts a particular conformation characterized by bringing together the 3'-end and the TPsiC loop. This is a first evidence for implication of RNA secondary structure rearrangement in the mechanism of mitochondrial import selectivity. Based on these data, a set of small RNA molecules with significantly improved efficiency of import into yeast and human mitochondria was constructed, opening the possibility of creating a new mitochondrial vector system able to target therapeutic oligoribonucleotides into deficient human mitochondria.
Figures


Similar articles
-
Induced tRNA import into human mitochondria: implication of a host aminoacyl-tRNA-synthetase.PLoS One. 2013 Jun 14;8(6):e66228. doi: 10.1371/journal.pone.0066228. Print 2013. PLoS One. 2013. PMID: 23799079 Free PMC article.
-
A Moonlighting Human Protein Is Involved in Mitochondrial Import of tRNA.Int J Mol Sci. 2015 Apr 24;16(5):9354-67. doi: 10.3390/ijms16059354. Int J Mol Sci. 2015. PMID: 25918939 Free PMC article.
-
Evidence for an adaptation mechanism of mitochondrial translation via tRNA import from the cytosol.Mol Cell. 2007 Jun 8;26(5):625-37. doi: 10.1016/j.molcel.2007.04.019. Mol Cell. 2007. PMID: 17560369
-
Mechanisms of tRNA import into yeast mitochondria: an overview.Biochimie. 1996;78(6):502-10. doi: 10.1016/0300-9084(96)84756-0. Biochimie. 1996. PMID: 8915539 Review.
-
Transfer RNA travels from the cytoplasm to organelles.Wiley Interdiscip Rev RNA. 2011 Nov-Dec;2(6):802-17. doi: 10.1002/wrna.93. Epub 2011 Jul 11. Wiley Interdiscip Rev RNA. 2011. PMID: 21976284 Free PMC article. Review.
Cited by
-
Single molecule tracking fluorescence microscopy in mitochondria reveals highly dynamic but confined movement of Tom40.Sci Rep. 2011;1:195. doi: 10.1038/srep00195. Epub 2011 Dec 14. Sci Rep. 2011. PMID: 22355710 Free PMC article.
-
Induced tRNA import into human mitochondria: implication of a host aminoacyl-tRNA-synthetase.PLoS One. 2013 Jun 14;8(6):e66228. doi: 10.1371/journal.pone.0066228. Print 2013. PLoS One. 2013. PMID: 23799079 Free PMC article.
-
Mitochondrial noncoding RNA transport.BMB Rep. 2017 Apr;50(4):164-174. doi: 10.5483/bmbrep.2017.50.4.013. BMB Rep. 2017. PMID: 28115039 Free PMC article. Review.
-
Correction of the consequences of mitochondrial 3243A>G mutation in the MT-TL1 gene causing the MELAS syndrome by tRNA import into mitochondria.Nucleic Acids Res. 2011 Oct;39(18):8173-86. doi: 10.1093/nar/gkr546. Epub 2011 Jun 30. Nucleic Acids Res. 2011. PMID: 21724600 Free PMC article.
-
tRNA biology in mitochondria.Int J Mol Sci. 2015 Feb 27;16(3):4518-59. doi: 10.3390/ijms16034518. Int J Mol Sci. 2015. PMID: 25734984 Free PMC article. Review.
References
-
- Brandina I, Graham J, Lemaitre-Guillier C, Entelis N, Krasheninnikov I, Sweetlove L, Tarassov I, Martin RP 2006. Enolase takes part in a macromolecular complex associated to mitochondria in yeast. Biochim Biophys Acta 1757: 1217–1228 - PubMed
-
- Conrad RC, Baskerville S, Ellington AD 1995. In vitro selection methodologies to probe RNA function and structure. Mol Divers 1: 69–78 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
