Residues in SRP9/14 essential for elongation arrest activity of the signal recognition particle define a positively charged functional domain on one side of the protein
- PMID: 20348448
- PMCID: PMC2856890
- DOI: 10.1261/rna.2040410
Residues in SRP9/14 essential for elongation arrest activity of the signal recognition particle define a positively charged functional domain on one side of the protein
Abstract
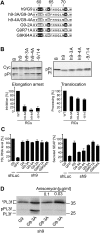
The signal recognition particle (SRP) is a ubiquitous cytoplasmic ribonucleoprotein complex required for the cotranslational targeting of proteins to the endoplasmic reticulum (ER). In eukaryotes, SRP has to arrest the elongation of the nascent chains during targeting to ensure efficient translocation of the preprotein, and this function of SRP is dependent on SRP9/14. Here we present the results of a mutational study on the human protein h9/14 that identified and characterized regions and single residues essential for elongation arrest activity. Effects of the mutations were assessed both in cell-free translation/translocation assays and in cultured mammalian cells. We identified two patches of basic amino acid residues that are essential for activity, whereas the internal loop of SRP14 was found to be dispensable. One patch of important basic residues comprises the previously identified basic pentapetide KRDKK, which can be substituted by four lysines without loss of function. The other patch includes three lysines in the solvent-accessible alpha2 of h9. All essential residues are located in proximity in SRP9/14 and their basic character suggests that they serve as a positively charged platform for interactions with ribosomal RNA. In addition, they can all be lysines consistent with the hypothesis that they recognize their target(s) via electrostatic contacts, most likely with the phosphate backbone, as opposed to contacts with specific bases.
Figures
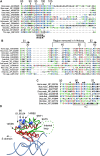
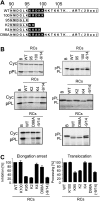
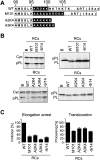
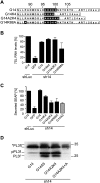
References
-
- Blau M, Mullapudi S, Becker T, Dudek J, Zimmermann R, Penczek PA, Beckmann R 2005. ERj1p uses a universal ribosomal adaptor site to coordinate the 80S ribosome at the membrane. Nat Struct Mol Biol 12: 1015–1016 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
