Urinary proteomics as a novel tool for biomarker discovery in kidney diseases
- PMID: 20349519
- PMCID: PMC2852539
- DOI: 10.1631/jzus.B0900327
Urinary proteomics as a novel tool for biomarker discovery in kidney diseases
Abstract
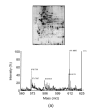
Urine has become one of the most attractive biofluids in clinical proteomics, for its procurement is easy and noninvasive and it contains sufficient proteins and peptides. Urinary proteomics has thus rapidly developed and has been extensively applied to biomarker discovery in clinical diseases, especially kidney diseases. In this review, we discuss two important aspects of urinary proteomics in detail, namely, sample preparation and proteomic technologies. In addition, data mining in urinary proteomics is also briefly introduced. At last, we present several successful examples on the application of urinary proteomics for biomarker discovery in kidney diseases, including diabetic nephropathy, IgA nephropathy, lupus nephritis, renal Fanconi syndrome, acute kidney injury, and renal allograft rejection.
Figures



References
-
- Dakna M, He Z, Yu WC, Mischak H, Kolch W. Technical, bioinformatical and statistical aspects of liquid chromatography-mass spectrometry (LC-MS) and capillary electrophoresis-mass spectrometry (CE-MS) based clinical proteomics: a critical assessment. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877(13):1250–1258. doi: 10.1016/j.jchromb.2008.10.048. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

