Large-scale analysis of full-length cDNAs from the tomato (Solanum lycopersicum) cultivar Micro-Tom, a reference system for the Solanaceae genomics
- PMID: 20350329
- PMCID: PMC2859864
- DOI: 10.1186/1471-2164-11-210
Large-scale analysis of full-length cDNAs from the tomato (Solanum lycopersicum) cultivar Micro-Tom, a reference system for the Solanaceae genomics
Abstract
Background: The Solanaceae family includes several economically important vegetable crops. The tomato (Solanum lycopersicum) is regarded as a model plant of the Solanaceae family. Recently, a number of tomato resources have been developed in parallel with the ongoing tomato genome sequencing project. In particular, a miniature cultivar, Micro-Tom, is regarded as a model system in tomato genomics, and a number of genomics resources in the Micro-Tom-background, such as ESTs and mutagenized lines, have been established by an international alliance.
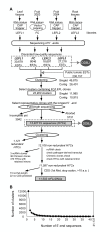
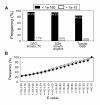
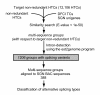
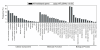
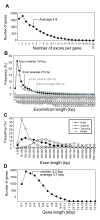
Results: To accelerate the progress in tomato genomics, we developed a collection of fully-sequenced 13,227 Micro-Tom full-length cDNAs. By checking redundant sequences, coding sequences, and chimeric sequences, a set of 11,502 non-redundant full-length cDNAs (nrFLcDNAs) was generated. Analysis of untranslated regions demonstrated that tomato has longer 5'- and 3'-untranslated regions than most other plants but rice. Classification of functions of proteins predicted from the coding sequences demonstrated that nrFLcDNAs covered a broad range of functions. A comparison of nrFLcDNAs with genes of sixteen plants facilitated the identification of tomato genes that are not found in other plants, most of which did not have known protein domains. Mapping of the nrFLcDNAs onto currently available tomato genome sequences facilitated prediction of exon-intron structure. Introns of tomato genes were longer than those of Arabidopsis and rice. According to a comparison of exon sequences between the nrFLcDNAs and the tomato genome sequences, the frequency of nucleotide mismatch in exons between Micro-Tom and the genome-sequencing cultivar (Heinz 1706) was estimated to be 0.061%.
Conclusion: The collection of Micro-Tom nrFLcDNAs generated in this study will serve as a valuable genomic tool for plant biologists to bridge the gap between basic and applied studies. The nrFLcDNA sequences will help annotation of the tomato whole-genome sequence and aid in tomato functional genomics and molecular breeding. Full-length cDNA sequences and their annotations are provided in the database KaFTom http://www.pgb.kazusa.or.jp/kaftom/ via the website of the National Bioresource Project Tomato http://tomato.nbrp.jp.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

