Molecular epidemiology, sequence types, and plasmid analyses of KPC-producing Klebsiella pneumoniae strains in Israel
- PMID: 20350950
- PMCID: PMC2897314
- DOI: 10.1128/AAC.01818-09
Molecular epidemiology, sequence types, and plasmid analyses of KPC-producing Klebsiella pneumoniae strains in Israel
Abstract
Sporadic isolates of carbapenem-resistant KPC-2-producing Klebsiella pneumoniae were isolated in Tel Aviv Medical Center during 2005 and 2006, parallel to the emergence of the KPC-3-producing K. pneumoniae sequence type 258 (ST 258). We aimed to study the molecular epidemiology of these isolates and to characterize their bla(KPC)-carrying plasmids and their origin. Ten isolates (8 KPC-2 and 2 KPC-3 producing) were studied. All isolates were extremely drug resistant. They possessed the bla(KPC) gene and varied in their additional beta-lactamase contents. The KPC-2-producing strains belonged to three different sequence types: ST 340 (n = 2), ST 277 (n = 2), and a novel sequence type, ST 376 (n = 4). Among KPC-3-producing strains, a single isolate (ST 327) different from ST 258 was identified, but both strains carried the same plasmid (pKpQIL). The KPC-2-encoding plasmids varied in size (45 to 95 kb) and differed among each of the STs. Two of the Klebsiella bla(KPC-2)-carrying plasmids were identical to plasmids from Escherichia coli, suggesting a common origin of these plasmids. These data indicate that KPC evolution in K. pneumoniae is related to rare events of interspecies spread of bla(KPC-2)-carrying plasmids from E. coli followed by limited clonal spread, whereas KPC-3 carriage in this species is related almost strictly to clonal expansion of ST 258 carrying pKpQIL.
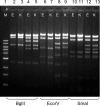
Figures


References
-
- Chmelnitsky, I., O. Hermesh, S. Navon-Venezia, J. Strahilevitz, and Y. Carmeli. 2009. First detection of aac (6′)-Ib-cr in KPC-producing Klebsiella pneumoniae isolates. J. Antimicrob. Chemother. 64:718-722. - PubMed
-
- Clinical and Laboratory Standards Institute. 2010. Performance standards for antimicrobial susceptibility testing; 20th informational supplement. M100-S20. Clinical and Laboratory Standards Institute, Wayne, PA.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

