Omp85 from the thermophilic cyanobacterium Thermosynechococcus elongatus differs from proteobacterial Omp85 in structure and domain composition
- PMID: 20351097
- PMCID: PMC2878562
- DOI: 10.1074/jbc.M110.112516
Omp85 from the thermophilic cyanobacterium Thermosynechococcus elongatus differs from proteobacterial Omp85 in structure and domain composition
Abstract
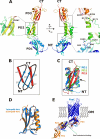
Omp85 proteins are essential proteins located in the bacterial outer membrane. They are involved in outer membrane biogenesis and assist outer membrane protein insertion and folding by an unknown mechanism. Homologous proteins exist in eukaryotes, where they mediate outer membrane assembly in organelles of endosymbiotic origin, the mitochondria and chloroplasts. We set out to explore the homologous relationship between cyanobacteria and chloroplasts, studying the Omp85 protein from the thermophilic cyanobacterium Thermosynechococcus elongatus. Using state-of-the art sequence analysis and clustering methods, we show how this protein is more closely related to its chloroplast homologue Toc75 than to proteobacterial Omp85, a finding supported by single channel conductance measurements. We have solved the structure of the periplasmic part of the protein to 1.97 A resolution, and we demonstrate that in contrast to Omp85 from Escherichia coli the protein has only three, not five, polypeptide transport-associated (POTRA) domains, which recognize substrates and generally interact with other proteins in bigger complexes. We model how these POTRA domains are attached to the outer membrane, based on the relationship of Omp85 to two-partner secretion system proteins, which we show and analyze. Finally, we discuss how Omp85 proteins with different numbers of POTRA domains evolved, and evolve to this day, to accomplish an increasing number of interactions with substrates and helper proteins.
Figures



References
-
- Voulhoux R., Bos M. P., Geurtsen J., Mols M., Tommassen J. (2003) Science 299, 262–265 - PubMed
-
- Wu T., Malinverni J., Ruiz N., Kim S., Silhavy T. J., Kahne D. (2005) Cell 121, 235–245 - PubMed
-
- Habib S. J., Waizenegger T., Lech M., Neupert W., Rapaport D. (2005) J. Biol. Chem. 280, 6434–6440 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

