The UBXN1 protein associates with autoubiquitinated forms of the BRCA1 tumor suppressor and inhibits its enzymatic function
- PMID: 20351172
- PMCID: PMC2876507
- DOI: 10.1128/MCB.01056-09
The UBXN1 protein associates with autoubiquitinated forms of the BRCA1 tumor suppressor and inhibits its enzymatic function
Abstract
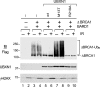
Although the BRCA1 tumor suppressor has been implicated in many cellular processes, the biochemical mechanisms by which it influences these diverse pathways are poorly understood. The only known enzymatic function of BRCA1 is the E3 ubiquitin ligase activity mediated by its highly conserved RING domain. In vivo, BRCA1 associates with the BARD1 polypeptide to form a heterodimeric BRCA1/BARD1 complex that catalyzes autoubiquitination of BRCA1 and trans ubiquitination of other protein substrates. In most cases, BRCA1-dependent ubiquitination generates polyubiquitin chains bearing an unconventional K6 linkage that does not appear to target proteins for proteasomal degradation. Since ubiquitin-dependent processes are usually mediated by cellular receptors with ubiquitin-binding motifs, we screened for proteins that specifically bind autoubiquitinated BRCA1. Here we report that the UBXN1 polypeptide, which contains a ubiquitin-associated (UBA) motif, recognizes autoubiquitinated BRCA1. This occurs through a bipartite interaction in which the UBA domain of UBXN1 binds K6-linked polyubiquitin chains conjugated to BRCA1 while the C-terminal sequences of UBXN1 bind the BRCA1/BARD1 heterodimer in a ubiquitin-independent fashion. Significantly, the E3 ligase activity of BRCA1/BARD1 is dramatically reduced in the presence of UBXN1, suggesting that UBXN1 regulates the enzymatic function of BRCA1 in a manner that is dependent on its ubiquitination status.
Figures










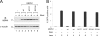
References
-
- Baer, R., and T. Ludwig. 2002. The BRCA1/BARD1 heterodimer, a tumor suppressor complex with ubiquitin E3 ligase activity. Curr. Opin. Genet. Dev. 12:86-91. - PubMed
-
- Chau, V., J. W. Tobias, A. Bachmair, D. Marriott, D. J. Ecker, D. K. Gonda, and A. Varshavsky. 1989. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 243:1576-1583. - PubMed
-
- Chen, A., F. E. Kleiman, J. L. Manley, T. Ouchi, and Z. Q. Pan. 2002. Auto-ubiquitination of the BRCA1/BARD1 RING ubiquitin ligase. J. Biol. Chem. 277:22085-22092. - PubMed
-
- Chen, Z. J., and L. J. Sun. 2009. Nonproteolytic functions of ubiquitin in cell signaling. Mol. Cell 33:275-286. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
