ErbB3/HER3 intracellular domain is competent to bind ATP and catalyze autophosphorylation
- PMID: 20351256
- PMCID: PMC2867849
- DOI: 10.1073/pnas.1002753107
ErbB3/HER3 intracellular domain is competent to bind ATP and catalyze autophosphorylation
Abstract
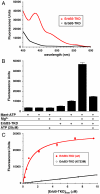
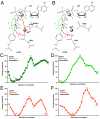
ErbB3/HER3 is one of four members of the human epidermal growth factor receptor (EGFR/HER) or ErbB receptor tyrosine kinase family. ErbB3 binds neuregulins via its extracellular region and signals primarily by heterodimerizing with ErbB2/HER2/Neu. A recently appreciated role for ErbB3 in resistance of tumor cells to EGFR/ErbB2-targeted therapeutics has made it a focus of attention. However, efforts to inactivate ErbB3 therapeutically in parallel with other ErbB receptors are challenging because its intracellular kinase domain is thought to be an inactive pseudokinase that lacks several key conserved (and catalytically important) residues-including the catalytic base aspartate. We report here that, despite these sequence alterations, ErbB3 retains sufficient kinase activity to robustly trans-autophosphorylate its intracellular region--although it is substantially less active than EGFR and does not phosphorylate exogenous peptides. The ErbB3 kinase domain binds ATP with a K(d) of approximately 1.1 microM. We describe a crystal structure of ErbB3 kinase bound to an ATP analogue, which resembles the inactive EGFR and ErbB4 kinase domains (but with a shortened alphaC-helix). Whereas mutations that destabilize this configuration activate EGFR and ErbB4 (and promote EGFR-dependent lung cancers), a similar mutation conversely inactivates ErbB3. Using quantum mechanics/molecular mechanics simulations, we delineate a reaction pathway for ErbB3-catalyzed phosphoryl transfer that does not require the conserved catalytic base and can be catalyzed by the "inactive-like" configuration observed crystallographically. These findings suggest that ErbB3 kinase activity within receptor dimers may be crucial for signaling and could represent an important therapeutic target.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

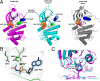
Comment in
-
Yet another "active" pseudokinase, Erb3.Proc Natl Acad Sci U S A. 2010 May 4;107(18):8047-8. doi: 10.1073/pnas.1003436107. Epub 2010 Apr 26. Proc Natl Acad Sci U S A. 2010. PMID: 20421461 Free PMC article. No abstract available.
References
-
- Hynes NE, MacDonald G. ErbB receptors and signaling pathways in cancer. Curr Opin Cell Biol. 2009;21:177–184. - PubMed
-
- Baselga J, Swain SM. Novel anticancer targets: Revisiting ERBB2 and discovering ERBB3. Nat Rev Cancer. 2009;9:463–475. - PubMed
-
- Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–1149. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

