Nucleocytoplasmic distribution and dynamics of the autophagosome marker EGFP-LC3
- PMID: 20352102
- PMCID: PMC2843706
- DOI: 10.1371/journal.pone.0009806
Nucleocytoplasmic distribution and dynamics of the autophagosome marker EGFP-LC3
Abstract
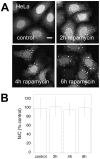
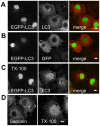
The process of autophagy involves the formation of autophagosomes, double-membrane structures that encapsulate cytosol. Microtubule-associated protein light chain 3 (LC3) was the first protein shown to specifically label autophagosomal membranes in mammalian cells, and subsequently EGFP-LC3 has become one of the most widely utilized reporters of autophagy. Although LC3 is currently thought to function primarily in the cytosol, the site of autophagosome formation, EGFP-LC3 often appears to be enriched in the nucleoplasm relative to the cytoplasm in published fluorescence images. However, the nuclear pool of EGFP-LC3 has not been specifically studied in previous reports, and mechanisms by which LC3 shuttles between the cytoplasm and nucleoplasm are currently unknown. In this study, we therefore investigated the regulation of the nucleo-cytoplasmic distribution of EGFP-LC3 in living cells. By quantitative fluorescence microscopy analysis, we demonstrate that soluble EGFP-LC3 is indeed enriched in the nucleus relative to the cytoplasm in two commonly studied cell lines, COS-7 and HeLa. Although LC3 contains a putative nuclear export signal (NES), inhibition of active nuclear export or mutation of the NES had no effect on the nucleo-cytoplasmic distribution of EGFP-LC3. Furthermore, FRAP analysis indicates that EGFP-LC3 undergoes limited passive nucleo-cytoplasmic transport under steady state conditions, and that the diffusional mobility of EGFP-LC3 was substantially slower in the nucleus and cytoplasm than predicted for a freely diffusing monomer. Induction of autophagy led to a visible decrease in levels of soluble EGFP-LC3 relative to autophagosome-bound protein, but had only modest effects on the nucleo-cytoplasmic ratio or diffusional mobility of the remaining soluble pools of EGFP-LC3. We conclude that the enrichment of soluble EGFP-LC3 in the nucleus is maintained independently of active nuclear export or induction of autophagy. Instead, incorporation of soluble EGFP-LC3 into large macromolecular complexes within both the cytoplasm and nucleus may prevent its rapid equilibrium between the two compartments.
Conflict of interest statement
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

