Abundant oligonucleotides common to most bacteria
- PMID: 20352124
- PMCID: PMC2843746
- DOI: 10.1371/journal.pone.0009841
Abundant oligonucleotides common to most bacteria
Abstract
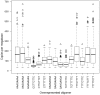
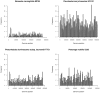
Background: Bacteria show a bias in their genomic oligonucleotide composition far beyond that dictated by G+C content. Patterns of over- and underrepresented oligonucleotides carry a phylogenetic signal and are thus diagnostic for individual species. Patterns of short oligomers have been investigated by multiple groups in large numbers of bacteria genomes. However, global distributions of the most highly overrepresented mid-sized oligomers have not been assessed across all prokaryotes to date. We surveyed overrepresented mid-length oligomers across all prokaryotes and normalised for base composition and embedded oligomers using zero and second order Markov models.
Principal findings: Here we report a presumably ancient set of oligomers conserved and overrepresented in nearly all branches of prokaryotic life, including Archaea. These oligomers are either adenine rich homopurines with one to three guanine nucleosides, or homopyridimines with one to four cytosine nucleosides. They do not show a consistent preference for coding or non-coding regions or aggregate in any coding frame, implying a role in DNA structure and as polypeptide binding sites. Structural parameters indicate these oligonucleotides to be an extreme and rigid form of B-DNA prone to forming triple stranded helices under common physiological conditions. Moreover, the narrow minor grooves of these structures are recognised by DNA binding and nucleoid associated proteins such as HU.
Conclusion: Homopurine and homopyrimidine oligomers exhibit distinct and unusual structural features and are present at high copy number in nearly all prokaryotic lineages. This fact suggests a non-neutral role of these oligonucleotides for bacterial genome organization that has been maintained throughout evolution.
Conflict of interest statement
Figures

Similar articles
-
Visualization of Pseudomonas genomic structure by abundant 8-14mer oligonucleotides.Environ Microbiol. 2009 May;11(5):1092-104. doi: 10.1111/j.1462-2920.2008.01839.x. Epub 2009 Jan 21. Environ Microbiol. 2009. PMID: 19161433
-
Sequence space coverage, entropy of genomes and the potential to detect non-human DNA in human samples.BMC Genomics. 2008 Oct 30;9:509. doi: 10.1186/1471-2164-9-509. BMC Genomics. 2008. PMID: 18973670 Free PMC article.
-
Investigations of oligonucleotide usage variance within and between prokaryotes.PLoS Comput Biol. 2008 Apr 18;4(4):e1000057. doi: 10.1371/journal.pcbi.1000057. PLoS Comput Biol. 2008. PMID: 18421372 Free PMC article.
-
[Gene identification in prokaryotic genomes using hidden Markov model].Tanpakushitsu Kakusan Koso. 1997 Dec;42(17 Suppl):2993-3000. Tanpakushitsu Kakusan Koso. 1997. PMID: 9455224 Review. Japanese. No abstract available.
-
The computational detection of functional nucleotide sequence motifs in the coding regions of organisms.Exp Biol Med (Maywood). 2008 Jun;233(6):665-73. doi: 10.3181/0704-MR-97. Epub 2008 Apr 11. Exp Biol Med (Maywood). 2008. PMID: 18408149 Review.
Cited by
-
Co-evolution of segregation guide DNA motifs and the FtsK translocase in bacteria: identification of the atypical Lactococcus lactis KOPS motif.Nucleic Acids Res. 2012 Jul;40(12):5535-45. doi: 10.1093/nar/gks171. Epub 2012 Feb 28. Nucleic Acids Res. 2012. PMID: 22373923 Free PMC article.
-
DistAMo: A Web-Based Tool to Characterize DNA-Motif Distribution on Bacterial Chromosomes.Front Microbiol. 2016 Mar 11;7:283. doi: 10.3389/fmicb.2016.00283. eCollection 2016. Front Microbiol. 2016. PMID: 27014208 Free PMC article.
References
-
- Dobrindt U, Hochhut B, Hentschel U, Hacker J. Genomic islands in pathogenic and environmental microorganisms. Nat Rev Microbiol. 2004;2:414–424. - PubMed
-
- Karlin S. Global dinucleotide signatures and analysis of genomic heterogeneity. Curr Opin Microbiol. 1998;1:598–610. - PubMed
-
- Davenport CF, Wiehlmann L, Reva ON, Tümmler B. Visualization of Pseudomonas genomic structure by abundant 8-14mer oligonucleotides. Environ Microbiol. 2009;11:1092–1104. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

