The yin and yang of chromatin spatial organization
- PMID: 20353545
- PMCID: PMC2864563
- DOI: 10.1186/gb-2010-11-3-204
The yin and yang of chromatin spatial organization
Abstract
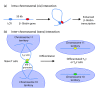
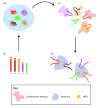
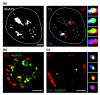
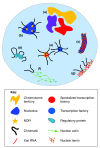
Spatial organization of the genome is non-random. Preferential chromatin interactions, both in cis and in trans and between transcriptionally active and silent regions, influence organization.
Figures
References
-
- Starck J, Sarkar R, Romana M, Bhargava A, Scarpa AL, Tanaka M, Chamberlain JW, Weissman SM, Forget BG. Developmental regulation of human gamma-globin and beta-globin genes in the absence of the locus-control region. Blood. 1994;84:1656–1665. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BB/E017460/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/B/0000C151/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/B/0000M723/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- G0800036/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources

