An insight into the sialome of Glossina morsitans morsitans
- PMID: 20353571
- PMCID: PMC2853526
- DOI: 10.1186/1471-2164-11-213
An insight into the sialome of Glossina morsitans morsitans
Abstract
Background: Blood feeding evolved independently in worms, arthropods and mammals. Among the adaptations to this peculiar diet, these animals developed an armament of salivary molecules that disarm their host's anti-bleeding defenses (hemostasis), inflammatory and immune reactions. Recent sialotranscriptome analyses (from the Greek sialo = saliva) of blood feeding insects and ticks have revealed that the saliva contains hundreds of polypeptides, many unique to their genus or family. Adult tsetse flies feed exclusively on vertebrate blood and are important vectors of human and animal diseases. Thus far, only limited information exists regarding the Glossina sialome, or any other fly belonging to the Hippoboscidae.
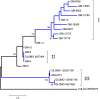
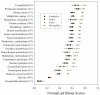
Results: As part of the effort to sequence the genome of Glossina morsitans morsitans, several organ specific, high quality normalized cDNA libraries have been constructed, from which over 20,000 ESTs from an adult salivary gland library were sequenced. These ESTs have been assembled using previously described ESTs from the fat body and midgut libraries of the same fly, thus totaling 62,251 ESTs, which have been assembled into 16,743 clusters (8,506 of which had one or more EST from the salivary gland library). Coding sequences were obtained for 2,509 novel proteins, 1,792 of which had at least one EST expressed in the salivary glands. Despite library normalization, 59 transcripts were overrepresented in the salivary library indicating high levels of expression. This work presents a detailed analysis of the salivary protein families identified. Protein expression was confirmed by 2D gel electrophoresis, enzymatic digestion and mass spectrometry. Concurrently, an initial attempt to determine the immunogenic properties of selected salivary proteins was undertaken.
Conclusions: The sialome of G. m. morsitans contains over 250 proteins that are possibly associated with blood feeding. This set includes alleles of previously described gene products, reveals new evidence that several salivary proteins are multigenic and identifies at least seven new polypeptide families unique to Glossina. Most of these proteins have no known function and thus, provide a discovery platform for the identification of novel pharmacologically active compounds, innovative vector-based vaccine targets, and immunological markers of vector exposure.
Figures
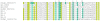





Similar articles
-
Characterization of genes expressed in the salivary glands of the tsetse fly, Glossina morsitans morsitans.Insect Mol Biol. 2001 Feb;10(1):69-76. doi: 10.1046/j.1365-2583.2001.00240.x. Insect Mol Biol. 2001. PMID: 11240638
-
Immunogenicity and Serological Cross-Reactivity of Saliva Proteins among Different Tsetse Species.PLoS Negl Trop Dis. 2015 Aug 27;9(8):e0004038. doi: 10.1371/journal.pntd.0004038. eCollection 2015. PLoS Negl Trop Dis. 2015. PMID: 26313460 Free PMC article.
-
The Glossina morsitans tsetse fly saliva: general characteristics and identification of novel salivary proteins.Insect Biochem Mol Biol. 2007 Oct;37(10):1075-85. doi: 10.1016/j.ibmb.2007.06.006. Epub 2007 Jun 20. Insect Biochem Mol Biol. 2007. PMID: 17785195
-
A case for a Glossina genome project.Trends Parasitol. 2005 Mar;21(3):107-11. doi: 10.1016/j.pt.2005.01.006. Trends Parasitol. 2005. PMID: 15734656 Review.
-
TickSialoFam (TSFam): A Database That Helps to Classify Tick Salivary Proteins, a Review on Tick Salivary Protein Function and Evolution, With Considerations on the Tick Sialome Switching Phenomenon.Front Cell Infect Microbiol. 2020 Jul 24;10:374. doi: 10.3389/fcimb.2020.00374. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 32850476 Free PMC article. Review.
Cited by
-
Sialome of a generalist lepidopteran herbivore: identification of transcripts and proteins from Helicoverpa armigera labial salivary glands.PLoS One. 2011;6(10):e26676. doi: 10.1371/journal.pone.0026676. Epub 2011 Oct 27. PLoS One. 2011. PMID: 22046331 Free PMC article.
-
Insight into the Salivary Gland Transcriptome of Lygus lineolaris (Palisot de Beauvois).PLoS One. 2016 Jan 20;11(1):e0147197. doi: 10.1371/journal.pone.0147197. eCollection 2016. PLoS One. 2016. PMID: 26789269 Free PMC article.
-
Tsetse salivary glycoproteins are modified with paucimannosidic N-glycans, are recognised by C-type lectins and bind to trypanosomes.PLoS Negl Trop Dis. 2021 Feb 2;15(2):e0009071. doi: 10.1371/journal.pntd.0009071. eCollection 2021 Feb. PLoS Negl Trop Dis. 2021. PMID: 33529215 Free PMC article.
-
Vaccines to combat the neglected tropical diseases.Immunol Rev. 2011 Jan;239(1):237-70. doi: 10.1111/j.1600-065X.2010.00976.x. Immunol Rev. 2011. PMID: 21198676 Free PMC article. Review.
-
Lysophosphatidylcholine: A Novel Modulator of Trypanosoma cruzi Transmission.J Parasitol Res. 2012;2012:625838. doi: 10.1155/2012/625838. Epub 2011 Nov 1. J Parasitol Res. 2012. PMID: 22132309 Free PMC article.
References
-
- Courtney GW, Pape T, Skevington JH, Sinclair BJ. In: Insect Biodiversity: Science and Society. Foottit RG, Adler PH, editor. Oxford: Blackwell Publishing; 2009. Biodiversity of Diptera; pp. 185–222. full_text.
-
- Grimaldi D, Engel M. Evolution of the insects. New York: Cambridge University Press; 2005.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

