F-box proteins FKF1 and LKP2 act in concert with ZEITLUPE to control Arabidopsis clock progression
- PMID: 20354196
- PMCID: PMC2861467
- DOI: 10.1105/tpc.109.072843
F-box proteins FKF1 and LKP2 act in concert with ZEITLUPE to control Arabidopsis clock progression
Abstract
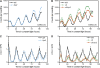
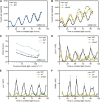
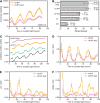
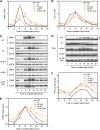
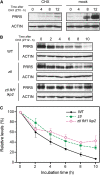
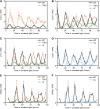
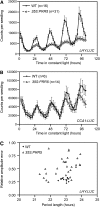
Regulation of protein turnover mediated by ZEITLUPE (ZTL) constitutes an important mechanism of the circadian clock in Arabidopsis thaliana. Here, we report that FLAVIN BINDING, KELCH REPEAT, F-BOX1 (FKF1) and LOV KELCH PROTEIN2 (LKP2) play similar roles to ZTL in the circadian clock when ZTL is absent. In contrast with subtle circadian clock defects in fkf1, the clock in ztl fkf1 has a considerably longer period than in ztl. In ztl fkf1 lkp2, several clock parameters were even more severely affected than in ztl fkf1. Although LATE ELONGATED HYPOCOTYL (LHY) and CIRCADIAN CLOCK ASSOCIATED1 (CCA1) expression levels are lower in ztl than in the wild type, introducing both fkf1 and lkp2 mutations into the ztl mutant dramatically diminished LHY expression without further affecting CCA1 expression. This demonstrates different contributions of ZTL, FKF1, and LKP2 in the regulation of LHY and CCA1 expression. In addition, FKF1 and LKP2 also interacted with TIMING OF CAB EXPRESSION1 (TOC1) and PSEUDO-RESPONSE REGULATOR5 (PRR5), and both proteins were further stabilized in ztl fkf1 and ztl fkf1 lkp2 compared with in ztl. Our results indicate that ZTL, FKF1, and LKP2 together regulate TOC1 and PRR5 degradation and are major contributors to determining the period of circadian oscillation and enhancing robustness.
Figures

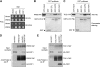
References
-
- Alabadi D., Oyama T., Yanovsky M.J., Harmon F.G., Más P., Kay S.A. (2001). Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293: 880–883 - PubMed
-
- Baudry A., Kay S.A. (2008). Clock control over plant gene expression. In Advances in Botanical Research, Vol. 48, Kader J.C., Delseny M., (San Diego, CA: Elsevier; ), pp. 70–105
-
- Briggs G.C., Osmont K.S., Shindo C., Sibout R., Hardtke C.S. (2006). Unequal genetic redundancies in Arabidopsis–A neglected phenomenon? Trends Plant Sci. 11: 492–498 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

