Spatial organization and signal transduction at intercellular junctions
- PMID: 20354536
- PMCID: PMC3693730
- DOI: 10.1038/nrm2883
Spatial organization and signal transduction at intercellular junctions
Abstract
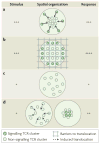
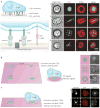
The coordinated organization of cell membrane receptors into diverse micrometre-scale spatial patterns is emerging as an important theme of intercellular signalling, as exemplified by immunological synapses. Key characteristics of these patterns are that they transcend direct protein-protein interactions, emerge transiently and modulate signal transduction. Such cooperativity over multiple length scales presents new and intriguing challenges for the study and ultimate understanding of cellular signalling. As a result, new experimental strategies have emerged to manipulate the spatial organization of molecules inside living cells. The resulting spatial mutations yield insights into the interweaving of the spatial, mechanical and chemical aspects of intercellular signalling.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
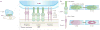



References
-
- Groves JT. Molecular organization and signal transduction at intermembrane junctions. Angew Chem Int Ed Engl. 2005;44:3524–3538. - PubMed
-
- Reich Z, et al. Ligand-specific oligomerization of T-cell receptor molecules. Nature. 1997;387:617–620. - PubMed
-
- Bray D, Levin MD, Morton-Firth CJ. Receptor clustering as a cellular mechanism to control sensitivity. Nature. 1998;393:85–88. - PubMed
-
- Maheshwari G, Brown G, Lauffenburger DA, Wells A, Griffith LG. Cell adhesion and motility depend on nanoscale RGD clustering. J Cell Sci. 2000;113:1677–1686. - PubMed
-
- Grakoui A, et al. The immunological synapse: a molecular machine controlling T cell activation. Science. 1999;285:221–227. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

