Linkage disequilibrium and age of HLA region SNPs in relation to classic HLA gene alleles within Europe
- PMID: 20354563
- PMCID: PMC2987379
- DOI: 10.1038/ejhg.2010.32
Linkage disequilibrium and age of HLA region SNPs in relation to classic HLA gene alleles within Europe
Abstract
The HLA region on chromosome 6 is gene-rich and under selective pressure because of the high proportion of immunity-related genes. Linkage disequilibrium (LD) patterns and allele frequencies in this region are highly differentiated across broad geographical populations, making it a region of interest for population genetics and immunity-related disease studies. We examined LD in this important region of the genome in six European populations using 166 putatively neutral SNPs and the classical HLA-A, -B and -C gene alleles. We found that the pattern of association between classic HLA gene alleles and SNPs implied that most of the SNPs predated the origin of classic HLA gene alleles. The SNPs most strongly associated with HLA gene alleles were in some cases highly predictive of the HLA allele carrier status (misclassification rates ranged from <1 to 27%) in independent populations using five or fewer SNPs, a much smaller number than tagSNP panels previously proposed and often with similar accuracy, showing that our approach may be a viable solution to designing new HLA prediction panels. To describe the LD within this region, we developed a new haplotype clustering method/software based on r(2), which may be more appropriate for use within regions of strong LD. Haplotype blocks created using this proposed method, as well as classic HLA gene alleles and SNPs, were predictive of a northern versus southern European population membership (misclassification error rates ranged from 0 to 23%, depending on which independent population was used for prediction), indicating that this region may be a rich source of ancestry informative markers.
Figures

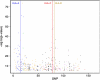
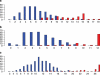

References
-
- Shen R, Fan JB, Campbell D, et al. High-throughput SNP genotyping on universal bead arrays. Mutat Res. 2005;573:70–82. - PubMed
-
- Tonks S, Marsh S, Bunce M, Bodmer JG. Molecular typing for HLA class I using ARMS-PCR: further development following the 12th International Histocompatibility Workshop. Tissue Antigens. 1999;53:175–183. - PubMed
-
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

