Structure of the ternary complex formed by a chemotaxis receptor signaling domain, the CheA histidine kinase, and the coupling protein CheW as determined by pulsed dipolar ESR spectroscopy
- PMID: 20355710
- PMCID: PMC2873776
- DOI: 10.1021/bi100055m
Structure of the ternary complex formed by a chemotaxis receptor signaling domain, the CheA histidine kinase, and the coupling protein CheW as determined by pulsed dipolar ESR spectroscopy
Abstract
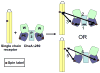
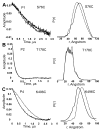
The signaling apparatus that controls bacterial chemotaxis is composed of a core complex containing chemoreceptors, the histidine autokinase CheA, and the coupling protein CheW. Site-specific spin labeling and pulsed dipolar ESR spectroscopy (PDS) have been applied to investigate the structure of a soluble ternary complex formed by Thermotoga maritima CheA (TmCheA), CheW, and receptor signaling domains. Thirty-five symmetric spin-label sites (SLSs) were engineered into the five domains of the CheA dimer and CheW to provide distance restraints within the CheA:CheW complex in the absence and presence of a soluble receptor that inhibits kinase activity (Tm14). Additional PDS restraints among spin-labeled CheA, CheW, and an engineered single-chain receptor labeled at six different sites allow docking of the receptor structure relative to the CheA:CheW complex. Disulfide cross-linking between selectively incorporated Cys residues finds two pairs of positions that provide further constraints within the ternary complex: one involving Tm14 and CheW and another involving Tm14 and CheA. The derived structure of the ternary complex indicates a primary site of interaction between CheW and Tm14 that agrees well with previous biochemical and genetic data for transmembrane chemoreceptors. The PDS distance distributions are most consistent with only one CheW directly engaging one dimeric Tm14. The CheA dimerization domain (P3) aligns roughly antiparallel to the receptor-conserved signaling tip but does not interact strongly with it. The angle of the receptor axis with respect to P3 and the CheW-binding P5 domains is bound by two limits differing by approximately 20 degrees . In one limit, Tm14 aligns roughly along P3 and may interact to some extent with the hinge region near the P3 hairpin loop. In the other limit, Tm14 tilts to interact with the P5 domain of the opposite subunit in an interface that mimics that observed with the P5 homologue CheW. The time domain ESR data can be simulated from the model only if orientational variability is introduced for the P5 and, especially, P3 domains. The Tm14 tip also binds beside one of the CheA kinase domains (P4); however, in both bound and unbound states, P4 samples a broad range of distributions that are only minimally affected by Tm14 binding. The CheA P1 domains that contain the substrate histidine are also broadly distributed in space under all conditions. In the context of the hexagonal lattice formed by trimeric transmembrane chemoreceptors, the PDS structure is best accommodated with the P3 domain in the center of a honeycomb edge.
Figures
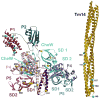
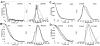
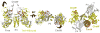
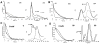

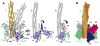
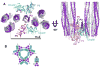
References
-
- Wadhams GH, Armitage JP. Making sense of it all: bacterial chemotaxis. Nat Rev Mol Cell Biol. 2004;5:1024–1037. - PubMed
-
- Parkinson JS, Kofoid EC. Communication modules in bacterial signaling proteins. Annu Rev Genet. 1992;26:71–112. - PubMed
-
- Sourjik V. Receptor clustering and signal processing in E. coli chemotaxis. Trends Microbiol. 2004;12:569–576. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

