Mouse Dfa is a repressor of TATA-box promoters and interacts with the Abt1 activator of basal transcription
- PMID: 20356838
- PMCID: PMC2878062
- DOI: 10.1074/jbc.M110.118638
Mouse Dfa is a repressor of TATA-box promoters and interacts with the Abt1 activator of basal transcription
Abstract
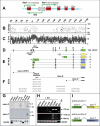
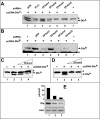
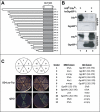
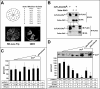
Our study of the mouse Ate1 arginyltransferase, a component of the N-end rule pathway, has shown that Ate1 pre-mRNA is produced from a bidirectional promoter that also expresses, in the opposite direction, a previously uncharacterized gene (Hu, R. G., Brower, C. S., Wang, H., Davydov, I. V., Sheng, J., Zhou, J., Kwon, Y. T., and Varshavsky, A. (2006) J. Biol. Chem. 281, 32559-32573). In this work, we began analyzing this gene, termed Dfa (divergent from Ate1). Mouse Dfa was found to be transcribed from both the bidirectional P(Ate1/Dfa) promoter and other nearby promoters. The resulting transcripts are alternatively spliced, yielding a complex set of Dfa mRNAs that are present largely, although not exclusively, in the testis. A specific Dfa mRNA encodes, via its 3'-terminal exon, a 217-residue protein termed Dfa(A). Other Dfa mRNAs also contain this exon. Dfa(A) is sequelogous (similar in sequence) to a region of the human/mouse HTEX4 protein, whose physiological function is unknown. We produced an affinity-purified antibody to recombinant mouse Dfa(A) that detected a 35-kDa protein in the mouse testis and in several cell lines. Experiments in which RNA interference was used to down-regulate Dfa indicated that the 35-kDa protein was indeed Dfa(A). Furthermore, Dfa(A) was present in the interchromatin granule clusters and was also found to bind to the Ggnbp1 gametogenetin-binding protein-1 and to the Abt1 activator of basal transcription that interacts with the TATA-binding protein. Given these results, RNA interference was used to probe the influence of Dfa levels in luciferase reporter assays. We found that Dfa(A) acts as a repressor of TATA-box transcriptional promoters.
Figures


Similar articles
-
Liat1, an arginyltransferase-binding protein whose evolution among primates involved changes in the numbers of its 10-residue repeats.Proc Natl Acad Sci U S A. 2014 Nov 18;111(46):E4936-45. doi: 10.1073/pnas.1419587111. Epub 2014 Nov 4. Proc Natl Acad Sci U S A. 2014. PMID: 25369936 Free PMC article.
-
ABT1-associated protein (ABTAP), a novel nuclear protein conserved from yeast to mammals, represses transcriptional activation by ABT1.J Cell Biochem. 2004 Nov 1;93(4):788-806. doi: 10.1002/jcb.20114. J Cell Biochem. 2004. PMID: 15660422
-
A novel TATA-binding protein-binding protein, ABT1, activates basal transcription and has a yeast homolog that is essential for growth.Mol Cell Biol. 2000 Feb;20(4):1407-18. doi: 10.1128/MCB.20.4.1407-1418.2000. Mol Cell Biol. 2000. PMID: 10648625 Free PMC article.
-
TEF-1 transrepression in BeWo cells is mediated through interactions with the TATA-binding protein, TBP.J Biol Chem. 1996 Apr 19;271(16):9510-8. doi: 10.1074/jbc.271.16.9510. J Biol Chem. 1996. PMID: 8621623
-
TRF2: TRansForming the view of general transcription factors.Transcription. 2015;6(1):1-6. doi: 10.1080/21541264.2015.1004980. Epub 2015 Jan 14. Transcription. 2015. PMID: 25588059 Free PMC article. Review.
Cited by
-
ATE1-Mediated Post-Translational Arginylation Is an Essential Regulator of Eukaryotic Cellular Homeostasis.ACS Chem Biol. 2020 Dec 18;15(12):3073-3085. doi: 10.1021/acschembio.0c00677. Epub 2020 Nov 23. ACS Chem Biol. 2020. PMID: 33228359 Free PMC article.
-
Liat1, an arginyltransferase-binding protein whose evolution among primates involved changes in the numbers of its 10-residue repeats.Proc Natl Acad Sci U S A. 2014 Nov 18;111(46):E4936-45. doi: 10.1073/pnas.1419587111. Epub 2014 Nov 4. Proc Natl Acad Sci U S A. 2014. PMID: 25369936 Free PMC article.
-
Characterization of cis-acting elements required for autorepression of the equine herpesvirus 1 IE gene.Virus Res. 2012 Apr;165(1):52-60. doi: 10.1016/j.virusres.2012.01.005. Epub 2012 Jan 14. Virus Res. 2012. PMID: 22265772 Free PMC article.
-
The N-end rule pathway and regulation by proteolysis.Protein Sci. 2011 Aug;20(8):1298-345. doi: 10.1002/pro.666. Protein Sci. 2011. PMID: 21633985 Free PMC article. Review.
-
TMT-based quantitative proteomics analysis reveals the attenuated replication mechanism of Newcastle disease virus caused by nuclear localization signal mutation in viral matrix protein.Virulence. 2020 Dec;11(1):607-635. doi: 10.1080/21505594.2020.1770482. Virulence. 2020. PMID: 32420802 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

