The recombination landscape of the zebra finch Taeniopygia guttata genome
- PMID: 20357052
- PMCID: PMC2847751
- DOI: 10.1101/gr.101410.109
The recombination landscape of the zebra finch Taeniopygia guttata genome
Abstract
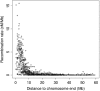
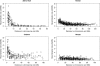
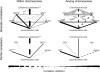
Understanding the causes and consequences of variation in the rate of recombination is essential since this parameter is considered to affect levels of genetic variability, the efficacy of selection, and the design of association and linkage mapping studies. However, there is limited knowledge about the factors governing recombination rate variation. We genotyped 1920 single nucleotide polymorphisms in a multigeneration pedigree of more than 1000 zebra finches (Taeniopygia guttata) to develop a genetic linkage map, and then we used these map data together with the recently available draft genome sequence of the zebra finch to estimate recombination rates in 1 Mb intervals across the genome. The average zebra finch recombination rate (1.5 cM/Mb) is higher than in humans, but significantly lower than in chicken. The local rates of recombination in chicken and zebra finch were only weakly correlated, demonstrating evolutionary turnover of the recombination landscape in birds. The distribution of recombination events was heavily biased toward ends of chromosomes, with a stronger telomere effect than so far seen in any organism. In fact, the recombination rate was as low as 0.1 cM/Mb in intervals up to 100 Mb long in the middle of the larger chromosomes. We found a positive correlation between recombination rate and GC content, as well as GC-rich sequence motifs. Levels of linkage disequilibrium (LD) were significantly higher in regions of low recombination, showing that heterogeneity in recombination rates have left a footprint on the genomic landscape of LD in zebra finch populations.
Figures



References
-
- Aerts J, Megens HJ, Veenendaal T, Ovcharenko I, Crooijmans R, Gordon L, Stubbs L, Groenen M. Extent of linkage disequilibrium in chicken. Cytogenet Genome Res. 2007;117:338–345. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
