Viral entry inhibitors targeted to the membrane site of action
- PMID: 20357085
- PMCID: PMC2903269
- DOI: 10.1128/JVI.00135-10
Viral entry inhibitors targeted to the membrane site of action
Abstract
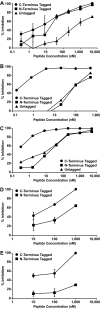
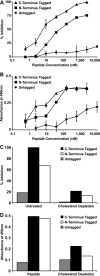
The fusion of enveloped viruses with the host cell is driven by specialized fusion proteins to initiate infection. The "class I" fusion proteins harbor two regions, typically two heptad repeat (HR) domains, which are central to the complex conformational changes leading to fusion: the first heptad repeat (HRN) is adjacent to the fusion peptide, while the second (HRC) immediately precedes the transmembrane domain. Peptides derived from the HR regions can inhibit fusion, and one HR peptide, T20 (enfuvirtide), is in clinical use for HIV-1. For paramyxoviruses, the activities of two membrane proteins, the receptor-binding protein (hemagglutinin-neuraminidase [HN] or G) and the fusion protein (F), initiate viral entry. The binding of HN or G to its receptor on a target cell triggers the activation of F, which then inserts into the target cell and mediates the membrane fusion that initiates infection. We have shown that for paramyxoviruses, the inhibitory efficacy of HR peptides is inversely proportional to the rate of F activation. For HIV-1, the antiviral potency of an HRC-derived peptide can be dramatically increased by targeting it to the membrane microdomains where fusion occurs, via the addition of a cholesterol group. We report here that for three paramyxoviruses-human parainfluenza virus type 3 (HPIV3), a major cause of lower respiratory tract diseases in infants, and the emerging zoonotic viruses Hendra virus (HeV) and Nipah virus (NiV), which cause lethal central nervous system diseases-the addition of cholesterol to a paramyxovirus HRC-derived peptide increased antiviral potency by 2 log units. Our data suggest that this enhanced activity is indeed the result of the targeting of the peptide to the plasma membrane, where fusion occurs. The cholesterol-tagged peptides on the cell surface create a protective antiviral shield, target the F protein directly at its site of action, and expand the potential utility of inhibitory peptides for paramyxoviruses.
Figures
Similar articles
-
Molecular determinants of antiviral potency of paramyxovirus entry inhibitors.J Virol. 2007 Oct;81(19):10567-74. doi: 10.1128/JVI.01181-07. Epub 2007 Jul 25. J Virol. 2007. PMID: 17652384 Free PMC article.
-
Inhibiting Human Parainfluenza Virus Infection by Preactivating the Cell Entry Mechanism.mBio. 2019 Feb 19;10(1):e02900-18. doi: 10.1128/mBio.02900-18. mBio. 2019. PMID: 30782664 Free PMC article.
-
Inhibition of Nipah virus infection in vivo: targeting an early stage of paramyxovirus fusion activation during viral entry.PLoS Pathog. 2010 Oct 28;6(10):e1001168. doi: 10.1371/journal.ppat.1001168. PLoS Pathog. 2010. PMID: 21060819 Free PMC article.
-
Detailed Molecular Biochemistry for Novel Therapeutic Design Against Nipah and Hendra Virus: A Systematic Review.Curr Mol Pharmacol. 2020;13(2):108-125. doi: 10.2174/1874467212666191023123732. Curr Mol Pharmacol. 2020. PMID: 31657692
-
Henipavirus mediated membrane fusion, virus entry and targeted therapeutics.Viruses. 2012 Feb;4(2):280-308. doi: 10.3390/v4020280. Epub 2012 Feb 13. Viruses. 2012. PMID: 22470837 Free PMC article. Review.
Cited by
-
A cholesterol tag at the N terminus of the relatively broad-spectrum fusion inhibitory peptide targets an earlier stage of fusion glycoprotein activation and increases the peptide's antiviral potency in vivo.J Virol. 2013 Aug;87(16):9223-32. doi: 10.1128/JVI.01153-13. Epub 2013 Jun 26. J Virol. 2013. PMID: 23804636 Free PMC article.
-
Engineering Protease-Resistant Peptides to Inhibit Human Parainfluenza Viral Respiratory Infection.J Am Chem Soc. 2021 Apr 21;143(15):5958-5966. doi: 10.1021/jacs.1c01565. Epub 2021 Apr 7. J Am Chem Soc. 2021. PMID: 33825470 Free PMC article.
-
Conformational and lipid bilayer-perturbing properties of Marburg virus GP2 segments containing the fusion loop and membrane-proximal external region/transmembrane domain.Heliyon. 2019 Dec 12;5(12):e03018. doi: 10.1016/j.heliyon.2019.e03018. eCollection 2019 Dec. Heliyon. 2019. PMID: 31890962 Free PMC article.
-
Microbial Metabolites: The Emerging Hotspot of Antiviral Compounds as Potential Candidates to Avert Viral Pandemic Alike COVID-19.Front Mol Biosci. 2021 Sep 7;8:732256. doi: 10.3389/fmolb.2021.732256. eCollection 2021. Front Mol Biosci. 2021. PMID: 34557521 Free PMC article. Review.
-
How a paramyxovirus fusion/entry complex adapts to escape a neutralizing antibody.Nat Commun. 2024 Oct 12;15(1):8831. doi: 10.1038/s41467-024-53082-y. Nat Commun. 2024. PMID: 39396053 Free PMC article.
References
-
- Baker, K. A., et al. 1999. Structural basis for paramyxovirus-mediated membrane fusion. Mol. Cell 3:309-319. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

