The Rab11 pathway is required for influenza A virus budding and filament formation
- PMID: 20357086
- PMCID: PMC2876627
- DOI: 10.1128/JVI.00307-10
The Rab11 pathway is required for influenza A virus budding and filament formation
Abstract
Influenza A virus buds through the apical plasma membrane, forming enveloped virus particles that can take the shape of pleomorphic spheres or vastly elongated filaments. For either type of virion, the factors responsible for separation of viral and cell membranes are not known. We find that cellular Rab11 (a small GTP-binding protein involved in endocytic recycling) and Rab11-family interacting protein 3 ([FIP3] which plays a role in membrane trafficking and regulation of actin dynamics) are both required to support the formation of filamentous virions, while Rab11 is additionally involved in the final budding step of spherical particles. Cells transfected with Rab11 GTP-cycling mutants or depleted of Rab11 or FIP3 content by small interfering RNA treatment lost the ability to form virus filaments. Depletion of Rab11 resulted in up to a 100-fold decrease in titer of spherical virus released from cells. Scanning electron microscopy of Rab11-depleted cells showed high densities of virus particles apparently stalled in the process of budding. Transmission electron microscopy of thin sections confirmed that Rab11 depletion resulted in significant numbers of abnormally formed virus particles that had failed to pinch off from the plasma membrane. Based on these findings, we see a clear role for a Rab11-mediated pathway in influenza virus morphogenesis and budding.
Figures

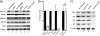
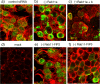


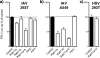
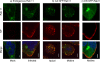



References
-
- Amorim, M. J., E. K. Read, R. M. Dalton, L. Medcalf, and P. Digard. 2007. Nuclear export of influenza A virus mRNAs requires ongoing RNA polymerase II activity. Traffic 8:1-11. - PubMed
-
- Bourmakina, S. V., and A. Garcia-Sastre. 2003. Reverse genetics studies on the filamentous morphology of influenza A virus. J. Gen. Virol. 84:517-527. - PubMed
-
- Bruce, E. A., L. Medcalf, C. M. Crump, S. L. Noton, A. D. Stuart, H. M. Wise, D. Elton, K. Bowers, and P. Digard. 2009. Budding of filamentous and non-filamentous influenza A virus occurs via a VPS4 and VPS28-independent pathway. Virology 390:268-278. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

