Interplay of RNA elements in the dengue virus 5' and 3' ends required for viral RNA replication
- PMID: 20357095
- PMCID: PMC2876622
- DOI: 10.1128/JVI.02042-09
Interplay of RNA elements in the dengue virus 5' and 3' ends required for viral RNA replication
Abstract
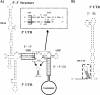
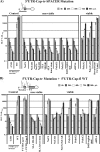
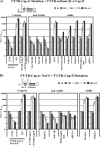
Dengue virus (DENV) is a member of the Flavivirus genus of positive-sense RNA viruses. DENV RNA replication requires cyclization of the viral genome mediated by two pairs of complementary sequences in the 5' and 3' ends, designated 5' and 3' cyclization sequences (5'-3' CS) and the 5' and 3' upstream of AUG region (5'-3' UAR). Here, we demonstrate that another stretch of six nucleotides in the 5' end is involved in DENV replication and possibly genome cyclization. This new sequence is located downstream of the AUG, designated the 5' downstream AUG region (5' DAR); the motif predicted to be complementary in the 3' end is termed the 3' DAR. In addition to the UAR, CS and DAR motifs, two other RNA elements are located at the 5' end of the viral RNA: the 5' stem-loop A (5' SLA) interacts with the viral RNA-dependent RNA polymerase and promotes RNA synthesis, and a stem-loop in the coding region named cHP is involved in translation start site selection as well as RNA replication. We analyzed the interplay of these 5' RNA elements in relation to RNA replication, and our data indicate that two separate functional units are formed; one consists of the SLA, and the other includes the UAR, DAR, cHP, and CS elements. The SLA must be located at the 5' end of the genome, whereas the position of the second unit is more flexible. We also show that the UAR, DAR, cHP, and CS must act in concert and therefore likely function together to form the tertiary RNA structure of the circularized DENV genome.
Figures






Similar articles
-
Structural and functional studies of the promoter element for dengue virus RNA replication.J Virol. 2009 Jan;83(2):993-1008. doi: 10.1128/JVI.01647-08. Epub 2008 Nov 12. J Virol. 2009. PMID: 19004935 Free PMC article.
-
Long-range RNA-RNA interactions circularize the dengue virus genome.J Virol. 2005 Jun;79(11):6631-43. doi: 10.1128/JVI.79.11.6631-6643.2005. J Virol. 2005. PMID: 15890901 Free PMC article.
-
Functional RNA elements in the dengue virus genome.Viruses. 2011 Sep;3(9):1739-56. doi: 10.3390/v3091739. Epub 2011 Sep 15. Viruses. 2011. PMID: 21994804 Free PMC article. Review.
-
RNA secondary structure in the coding region of dengue virus type 2 directs translation start codon selection and is required for viral replication.J Virol. 2006 Mar;80(5):2170-82. doi: 10.1128/JVI.80.5.2170-2182.2006. J Virol. 2006. PMID: 16474125 Free PMC article.
-
Structural and functional analysis of dengue virus RNA.Novartis Found Symp. 2006;277:120-32; discussion 132-5, 251-3. Novartis Found Symp. 2006. PMID: 17319158 Review.
Cited by
-
The RNA secondary structural variation in the cyclization elements of the dengue genome and the possible implications in pathogenicity.Virusdisease. 2020 Sep;31(3):299-307. doi: 10.1007/s13337-020-00615-w. Epub 2020 Jul 30. Virusdisease. 2020. PMID: 32904896 Free PMC article.
-
Dengue virus 2 capsid protein chaperones the strand displacement of 5'-3' cyclization sequences.Nucleic Acids Res. 2021 Jun 4;49(10):5832-5844. doi: 10.1093/nar/gkab379. Nucleic Acids Res. 2021. PMID: 34037793 Free PMC article.
-
Structural complexity of Dengue virus untranslated regions: cis-acting RNA motifs and pseudoknot interactions modulating functionality of the viral genome.Nucleic Acids Res. 2013 May;41(9):5075-89. doi: 10.1093/nar/gkt203. Epub 2013 Mar 26. Nucleic Acids Res. 2013. PMID: 23531545 Free PMC article.
-
Viral RNA switch mediates the dynamic control of flavivirus replicase recruitment by genome cyclization.Elife. 2016 Oct 1;5:e17636. doi: 10.7554/eLife.17636. Elife. 2016. PMID: 27692070 Free PMC article.
-
RNA structures required for production of subgenomic flavivirus RNA.J Virol. 2010 Nov;84(21):11407-17. doi: 10.1128/JVI.01159-10. Epub 2010 Aug 18. J Virol. 2010. PMID: 20719943 Free PMC article.
References
-
- Alvarez, D. E., C. V. Filomatori, and A. V. Gamarnik. 2008. Functional analysis of dengue virus cyclization sequences located at the 5′ and 3′UTRs. Virology 375:223-235. - PubMed
-
- Alvarez, D. E., A. L. Lella Ezcurra, S. Fucito, and A. V. Gamarnik. 2005. Role of RNA structures present at the 3′UTR of dengue virus on translation, RNA synthesis, and viral replication. Virology 339:200-212. - PubMed
-
- Alvarez, D. E., M. F. Lodeiro, C. V. Filomatori, S. Fucito, J. A. Mondotte, and A. V. Gamarnik. 2006. Structural and functional analysis of dengue virus RNA. Novartis Found. Symp. 277:120-132. - PubMed
-
- Astier-Gin, T., P. Bellecave, S. Litvak, and M. Ventura. 2005. Template requirements and binding of hepatitis C virus NS5B polymerase during in vitro RNA synthesis from the 3′-end of virus minus-strand RNA. FEBS J. 272:3872-3886. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

