Identification of a domain of the baculovirus Autographa californica multiple nucleopolyhedrovirus single-strand DNA-binding protein LEF-3 essential for viral DNA replication
- PMID: 20357098
- PMCID: PMC2876648
- DOI: 10.1128/JVI.00115-10
Identification of a domain of the baculovirus Autographa californica multiple nucleopolyhedrovirus single-strand DNA-binding protein LEF-3 essential for viral DNA replication
Abstract
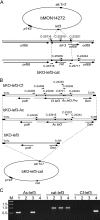
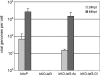
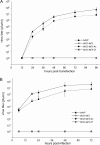
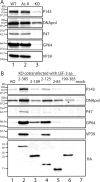
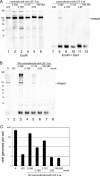
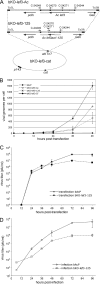
Autographa californica multiple nucleopolyhedrovirus (AcMNPV) lef-3 is one of nine genes required for viral DNA replication in transient assays. LEF-3 is predicted to contain several domains related to its functions, including nuclear localization, single-strand DNA binding, oligomerization, interaction with P143 helicase, and interaction with a viral alkaline nuclease. To investigate the essential nature of LEF-3 and the roles it may play during baculovirus DNA replication, a lef-3 null bacmid (bKO-lef3) was constructed in Escherichia coli and characterized in Sf21 cells. The results showed that AcMNPV lef-3 is essential for DNA replication, budded virus production, and late gene expression in vivo. Cells transfected with the lef-3 knockout bacmid produced low levels of early proteins (P143, DNA polymerase, and early GP64) and no late proteins (P47, VP39, or late GP64). To investigate the functional role of domains within the LEF-3 open reading frame in the presence of the whole viral genome, plasmids expressing various LEF-3 truncations were transfected into Sf21 cells together with bKO-lef3 DNA. The results showed that expression of AcMNPV LEF-3 amino acids 1 to 125 was sufficient to stimulate viral DNA replication and to support late gene expression. Expression of Choristoneura fumiferana MNPV lef-3 did not rescue any LEF-3 functions. The construction of a LEF-3 amino acid 1 to 125 rescue bacmid revealed that this region of LEF-3, when expressed in the presence of the rest of the viral genome, stimulated viral DNA replication and late and very late protein expression, as well as budded virus production.
Figures
Similar articles
-
Autographa californica multiple nucleopolyhedrovirus DNA polymerase C terminus is required for nuclear localization and viral DNA replication.J Virol. 2014 Sep;88(18):10918-33. doi: 10.1128/JVI.01167-14. Epub 2014 Jul 9. J Virol. 2014. PMID: 25008932 Free PMC article.
-
Choristoneura fumiferana multiple nucleopolyhedrovirus LEF-3-P143 complex can complement DNA replication and budded virus in an AcMNPV LEF-3-P143 double knockout bacmid.J Gen Virol. 2012 Feb;93(Pt 2):383-388. doi: 10.1099/vir.0.036699-0. Epub 2011 Nov 2. J Gen Virol. 2012. PMID: 22049089
-
Characterization of a baculovirus lacking the DBP (DNA-binding protein) gene.Virology. 2007 Aug 1;364(2):475-85. doi: 10.1016/j.virol.2007.03.024. Epub 2007 Apr 20. Virology. 2007. PMID: 17449080 Free PMC article.
-
me53 encoded by Autographa californica multiple nucleopolyhedrovirus: from mechanism to function.Virus Genes. 2023 Apr;59(2):188-194. doi: 10.1007/s11262-022-01943-3. Epub 2022 Oct 13. Virus Genes. 2023. PMID: 36229721 Review.
-
Baculovirus dual-phase infection strategy and biotechnological applications: A structural and regulatory review.Biotechnol Adv. 2025 Oct;83:108627. doi: 10.1016/j.biotechadv.2025.108627. Epub 2025 Jun 15. Biotechnol Adv. 2025. PMID: 40527357 Review.
Cited by
-
Analysis of the autographa californica multiple nucleopolyhedrovirus overlapping gene pair lef3 and ac68 reveals that AC68 is a per os infectivity factor and that LEF3 is critical, but not essential, for virus replication.J Virol. 2012 Apr;86(7):3985-94. doi: 10.1128/JVI.06849-11. Epub 2012 Jan 25. J Virol. 2012. PMID: 22278232 Free PMC article.
-
The logic of DNA replication in double-stranded DNA viruses: insights from global analysis of viral genomes.Nucleic Acids Res. 2016 Jun 2;44(10):4551-64. doi: 10.1093/nar/gkw322. Epub 2016 Apr 25. Nucleic Acids Res. 2016. PMID: 27112572 Free PMC article.
-
Use of bacterial artificial chromosomes in baculovirus research and recombinant protein expression: current trends and future perspectives.ISRN Microbiol. 2012 Sep 12;2012:628797. doi: 10.5402/2012/628797. Print 2012. ISRN Microbiol. 2012. PMID: 23762754 Free PMC article.
-
Global Analysis of Baculovirus Autographa californica Multiple Nucleopolyhedrovirus Gene Expression in the Midgut of the Lepidopteran Host Trichoplusia ni.J Virol. 2018 Nov 12;92(23):e01277-18. doi: 10.1128/JVI.01277-18. Print 2018 Dec 1. J Virol. 2018. PMID: 30209166 Free PMC article.
-
Autographa californica multiple nucleopolyhedrovirus DNA polymerase C terminus is required for nuclear localization and viral DNA replication.J Virol. 2014 Sep;88(18):10918-33. doi: 10.1128/JVI.01167-14. Epub 2014 Jul 9. J Virol. 2014. PMID: 25008932 Free PMC article.
References
-
- Au, V., M. Yu, and E. B. Carstens. 2009. Characterization of a baculovirus nuclear localization signal domain in the late expression factor 3 protein. Virology 385:209-217. - PubMed
-
- Blissard, G. W., and G. F. Rohrmann. 1989. Location, sequence, transcriptional mapping, and temporal expression of the gp64 envelope glycoprotein gene of the Orgyia pseudotsugata multicapsid nuclear polyhedrosis virus. Virology 170:537-555. - PubMed
-
- Carstens, E. B. 2009. AcMNPV as a model for baculovirus DNA replication. Virol. Sin. 24:243-267.
-
- Carstens, E. B., and Y. Wu. 2007. No single homologous repeat region is essential for DNA replication of the baculovirus Autographa californica multiple nucleopolyhedrovirus. J. Gen. Virol. 88:114-122. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

