Conformational changes in the capsid of a calicivirus upon interaction with its functional receptor
- PMID: 20357100
- PMCID: PMC2876613
- DOI: 10.1128/JVI.02371-09
Conformational changes in the capsid of a calicivirus upon interaction with its functional receptor
Abstract
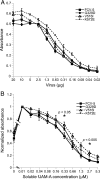
Nonenveloped viral capsids are metastable structures that undergo conformational changes during virus entry that lead to interactions of the capsid or capsid fragments with the cell membrane. For members of the Caliciviridae, neither the nature of these structural changes in the capsid nor the factor(s) responsible for inducing these changes is known. Feline junctional adhesion molecule A (fJAM-A) mediates the attachment and infectious viral entry of feline calicivirus (FCV). Here, we show that the infectivity of some FCV isolates is neutralized following incubation with the soluble receptor at 37 degrees C. We used this property to select mutants resistant to preincubation with the soluble receptor. We isolated and sequenced 24 soluble receptor-resistant (srr) mutants and characterized the growth properties and receptor-binding activities of eight mutants. The location of the mutations within the capsid structure of FCV was mapped using a new 3.6-A structure of native FCV. The srr mutations mapped to the surface of the P2 domain were buried at the protruding domain dimer interface or were present in inaccessible regions of the capsid protein. Coupled with data showing that both the parental FCV and the srr mutants underwent increases in hydrophobicity upon incubation with the soluble receptor at 37 degrees C, these findings indicate that FCV likely undergoes conformational change upon interaction with its receptor. Changes in FCV capsid conformation following its interaction with fJAM-A may be important for subsequent interactions of the capsid with cellular membranes, membrane penetration, and genome delivery.
Figures



Similar articles
-
The cryo-electron microscopy structure of feline calicivirus bound to junctional adhesion molecule A at 9-angstrom resolution reveals receptor-induced flexibility and two distinct conformational changes in the capsid protein VP1.J Virol. 2011 Nov;85(21):11381-90. doi: 10.1128/JVI.05621-11. Epub 2011 Aug 24. J Virol. 2011. PMID: 21865392 Free PMC article.
-
Conserved Surface Residues on the Feline Calicivirus Capsid Are Essential for Interaction with Its Receptor Feline Junctional Adhesion Molecule A (fJAM-A).J Virol. 2018 Mar 28;92(8):e00035-18. doi: 10.1128/JVI.00035-18. Print 2018 Apr 15. J Virol. 2018. PMID: 29386293 Free PMC article.
-
Identification of regions and residues in feline junctional adhesion molecule required for feline calicivirus binding and infection.J Virol. 2007 Dec;81(24):13608-21. doi: 10.1128/JVI.01509-07. Epub 2007 Oct 3. J Virol. 2007. PMID: 17913818 Free PMC article.
-
Structural insights into calicivirus attachment and uncoating.J Virol. 2008 Aug;82(16):8051-8. doi: 10.1128/JVI.00550-08. Epub 2008 Jun 11. J Virol. 2008. PMID: 18550656 Free PMC article.
-
VP2 mediates the release of the feline calicivirus RNA genome by puncturing the endosome membrane of infected cells.J Virol. 2024 May 14;98(5):e0035024. doi: 10.1128/jvi.00350-24. Epub 2024 Apr 9. J Virol. 2024. PMID: 38591900 Free PMC article.
Cited by
-
Conformational Flexibility in Capsids Encoded by the Caliciviridae.Viruses. 2024 Nov 26;16(12):1835. doi: 10.3390/v16121835. Viruses. 2024. PMID: 39772145 Free PMC article. Review.
-
The cryo-electron microscopy structure of feline calicivirus bound to junctional adhesion molecule A at 9-angstrom resolution reveals receptor-induced flexibility and two distinct conformational changes in the capsid protein VP1.J Virol. 2011 Nov;85(21):11381-90. doi: 10.1128/JVI.05621-11. Epub 2011 Aug 24. J Virol. 2011. PMID: 21865392 Free PMC article.
-
Efficacy of EPA-registered disinfectants against two human norovirus surrogates and Clostridioides difficile endospores.J Appl Microbiol. 2022 Jun;132(6):4289-4299. doi: 10.1111/jam.15524. Epub 2022 Mar 17. J Appl Microbiol. 2022. PMID: 35279925 Free PMC article.
-
Cryo-EM structure of a novel calicivirus, Tulane virus.PLoS One. 2013;8(3):e59817. doi: 10.1371/journal.pone.0059817. Epub 2013 Mar 22. PLoS One. 2013. PMID: 23533651 Free PMC article.
-
Conserved Surface Residues on the Feline Calicivirus Capsid Are Essential for Interaction with Its Receptor Feline Junctional Adhesion Molecule A (fJAM-A).J Virol. 2018 Mar 28;92(8):e00035-18. doi: 10.1128/JVI.00035-18. Print 2018 Apr 15. J Virol. 2018. PMID: 29386293 Free PMC article.
References
-
- Brünger, A. T. 1992. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature 355:472-475. - PubMed
-
- Brünger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54:905-921. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

