Loss of GluN2B-containing NMDA receptors in CA1 hippocampus and cortex impairs long-term depression, reduces dendritic spine density, and disrupts learning
- PMID: 20357110
- PMCID: PMC2869199
- DOI: 10.1523/JNEUROSCI.0640-10.2010
Loss of GluN2B-containing NMDA receptors in CA1 hippocampus and cortex impairs long-term depression, reduces dendritic spine density, and disrupts learning
Abstract
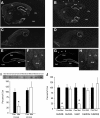
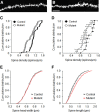
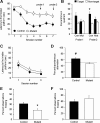
NMDA receptors (NMDARs) are key mediators of certain forms of synaptic plasticity and learning. NMDAR complexes are heteromers composed of an obligatory GluN1 subunit and one or more GluN2 (GluN2A-GluN2D) subunits. Different subunits confer distinct physiological and molecular properties to NMDARs, but their contribution to synaptic plasticity and learning in the adult brain remains uncertain. Here, we generated mice lacking GluN2B in pyramidal neurons of cortex and CA1 subregion of hippocampus. We found that hippocampal principal neurons of adult GluN2B mutants had faster decaying NMDAR-mediated EPSCs than nonmutant controls and were insensitive to GluN2B but not NMDAR antagonism. A subsaturating form of hippocampal long-term potentiation (LTP) was impaired in the mutants, whereas a saturating form of LTP was intact. An NMDAR-dependent form of long-term depression (LTD) produced by low-frequency stimulation combined with glutamate transporter inhibition was abolished in the mutants. Additionally, mutants exhibited decreased dendritic spine density in CA1 hippocampal neurons compared with controls. On multiple assays for corticohippocampal-mediated learning and memory (hidden platform Morris water maze, T-maze spontaneous alternation, and pavlovian trace fear conditioning), mutants were impaired. These data further demonstrate the importance of GluN2B for synaptic plasticity in the adult hippocampus and suggest a particularly critical role in LTD, at least the form studied here. The finding that loss of GluN2B was sufficient to cause learning deficits illustrates the contribution of GluN2B-mediated forms of plasticity to memory formation, with implications for elucidating NMDAR-related dysfunction in disease-related cognitive impairment.
Figures


References
-
- Akashi K, Kakizaki T, Kamiya H, Fukaya M, Yamasaki M, Abe M, Natsume R, Watanabe M, Sakimura K. NMDA receptor GluN2B (GluRε2/NR2B) subunit is crucial for channel function, postsynaptic macromolecular organization, and actin cytoskeleton at hippocampal CA3 synapses. J Neurosci. 2009;29:10869–10882. - PMC - PubMed
-
- Alvarez VA, Sabatini BL. Anatomical and physiological plasticity of dendritic spines. Annu Rev Neurosci. 2007;30:79–97. - PubMed
-
- Bannerman DM, Rawlins JN, Good MA. The drugs don't work-or do they? Pharmacological and transgenic studies of the contribution of NMDA and GluR-A-containing AMPA receptors to hippocampal-dependent memory. Psychopharmacology (Berl) 2006;188:552–566. - PubMed
-
- Bannerman DM, Niewoehner B, Lyon L, Romberg C, Schmitt WB, Taylor A, Sanderson DJ, Cottam J, Sprengel R, Seeburg PH, Köhr G, Rawlins JN. NMDA receptor subunit NR2A is required for rapidly acquired spatial working memory but not incremental spatial reference memory. J Neurosci. 2008;28:3623–3630. - PMC - PubMed
-
- Barria A, Malinow R. NMDA receptor subunit composition controls synaptic plasticity by regulating binding to CaMKII. Neuron. 2005;48:289–301. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
