Integrating ELF4 into the circadian system through combined structural and functional studies
- PMID: 20357892
- PMCID: PMC2801535
- DOI: 10.2976/1.3218766
Integrating ELF4 into the circadian system through combined structural and functional studies
Abstract
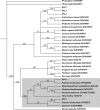
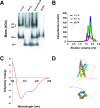
The circadian clock is a timekeeping mechanism that enables anticipation of daily environmental changes. In the plant Arabidopsis thaliana, the circadian system is a multiloop series of interlocked transcription-translation feedbacks. Several genes have been arranged in these oscillation loops, but the position of the core-clock gene ELF4 in this network was previously undetermined. ELF4 lacks sequence similarity to known domains, and functional homologs have not yet been identified. Here we show that ELF4 is functionally conserved within a subclade of related sequences, and forms an alpha-helical homodimer with a likely electrostatic interface that could be structurally modeled. We support this hypothesis by expression analysis of new elf4 hypomorphic alleles. These weak mutants were found to have expression level phenotypes of both morning and evening clock genes, implicating multiple entry points of ELF4 within the multiloop network. This could be mathematically modeled. Furthermore, morning-expression defects were particular to some elf4 alleles, suggesting predominant ELF4 action just preceding dawn. We provide a new hypothesis about ELF4 in the oscillator-it acts as a homodimer to integrate two arms of the circadian clock.
Figures






References
-
- Altschul, S F, Gish, W, Miller, W, Myers, E W, and Lipman, D J (1990). “Basic local alignment search tool.” J. Mol. Biol. JMOBAK 215, 403–410. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
