A novel intronic peroxisome proliferator-activated receptor gamma enhancer in the uncoupling protein (UCP) 3 gene as a regulator of both UCP2 and -3 expression in adipocytes
- PMID: 20360005
- PMCID: PMC2878494
- DOI: 10.1074/jbc.M110.120584
A novel intronic peroxisome proliferator-activated receptor gamma enhancer in the uncoupling protein (UCP) 3 gene as a regulator of both UCP2 and -3 expression in adipocytes
Abstract
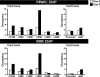
Uncoupling Proteins (UCPs) are integral ion channels residing in the inner mitochondrial membrane. UCP2 is ubiquitously expressed, while UCP3 is found primarily in muscles and adipose tissue. Although the exact molecular mechanism of action is controversial, it is generally agreed that both homologues function to facilitate mitochondrial fatty acid oxidation. UCP2 and -3 expression is activated by the peroxisome proliferator-activated receptors (PPARs), but so far no PPAR response element has been reported in the vicinity of the Ucp2 and Ucp3 genes. Using genome-wide profiling of PPARgamma occupancy in 3T3-L1 adipocytes we demonstrate that PPARgamma associates with three chromosomal regions in the vicinity of the Ucp3 locus and weakly with a site in intron 1 of the Ucp2 gene. These sites are isolated from the nearest neighboring sites by >900 kb. The most prominent PPARgamma binding site in the Ucp2 and Ucp3 loci is located in intron 1 of the Ucp3 gene and is the only site that facilitates PPARgamma transactivation of a heterologous promoter. This site furthermore transactivates the endogenous Ucp3 promoter, and using chromatin conformation capture we show that it loops out to specifically interact with the Ucp2 promoter and intron 1. Our data indicate that PPARgamma transactivation of both UCP2 and -3 is mediated through this novel enhancer in Ucp3 intron 1.
Figures
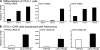

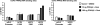
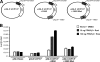

Similar articles
-
Mechanism of ubiquitous expression of mouse uncoupling protein 2 mRNA: control by cis-acting DNA element in 5'-flanking region.Biochem J. 1999 Jun 1;340 ( Pt 2)(Pt 2):397-404. Biochem J. 1999. PMID: 10333481 Free PMC article.
-
A novel SP1/SP3 dependent intronic enhancer governing transcription of the UCP3 gene in brown adipocytes.PLoS One. 2013 Dec 31;8(12):e83426. doi: 10.1371/journal.pone.0083426. eCollection 2013. PLoS One. 2013. PMID: 24391766 Free PMC article.
-
Genomic organization and mutational analysis of the human UCP2 gene, a prime candidate gene for human obesity.J Recept Signal Transduct Res. 1999 Jan-Jul;19(1-4):229-44. doi: 10.3109/10799899909036648. J Recept Signal Transduct Res. 1999. PMID: 10071761
-
Uncoupling Proteins in Striated Muscle Tissue: Known Facts and Open Questions.Antioxid Redox Signal. 2022 Aug;37(4-6):324-335. doi: 10.1089/ars.2021.0258. Epub 2022 Apr 18. Antioxid Redox Signal. 2022. PMID: 35044239 Review.
-
Links between fatty acids and expression of UCP2 and UCP3 mRNAs.FEBS Lett. 2004 Jun 18;568(1-3):4-9. doi: 10.1016/j.febslet.2004.05.011. FEBS Lett. 2004. PMID: 15196910 Review.
Cited by
-
PPARγ lipodystrophy mutants reveal intermolecular interactions required for enhancer activation.Nat Commun. 2022 Nov 19;13(1):7090. doi: 10.1038/s41467-022-34766-9. Nat Commun. 2022. PMID: 36402763 Free PMC article.
-
PPARγ and the global map of adipogenesis and beyond.Trends Endocrinol Metab. 2014 Jun;25(6):293-302. doi: 10.1016/j.tem.2014.04.001. Epub 2014 Apr 29. Trends Endocrinol Metab. 2014. PMID: 24793638 Free PMC article. Review.
-
Uncoupling Protein 2: A Key Player and a Potential Therapeutic Target in Vascular Diseases.Oxid Med Cell Longev. 2017;2017:7348372. doi: 10.1155/2017/7348372. Epub 2017 Oct 15. Oxid Med Cell Longev. 2017. PMID: 29163755 Free PMC article. Review.
-
UCP2, a mitochondrial protein regulated at multiple levels.Cell Mol Life Sci. 2014 Apr;71(7):1171-90. doi: 10.1007/s00018-013-1407-0. Epub 2013 Jun 27. Cell Mol Life Sci. 2014. PMID: 23807210 Free PMC article. Review.
-
Honokiol Ameliorates Post-Myocardial Infarction Heart Failure Through Ucp3-Mediated Reactive Oxygen Species Inhibition.Front Pharmacol. 2022 Feb 21;13:811682. doi: 10.3389/fphar.2022.811682. eCollection 2022. Front Pharmacol. 2022. PMID: 35264952 Free PMC article.
References
-
- Heaton G. M., Wagenvoord R. J., Kemp A., Jr., Nicholls D. G. (1978) Eur. J. Biochem. 82, 515–521 - PubMed
-
- Cannon B., Nedergaard J. (2004) Physiol. Rev. 84, 277–359 - PubMed
-
- Fleury C., Neverova M., Collins S., Raimbault S., Champigny O., Levi-Meyrueis C., Bouillaud F., Seldin M. F., Surwit R. S., Ricquier D., Warden C. H. (1997) Nat. Genet. 15, 269–272 - PubMed
-
- Boss O., Samec S., Paoloni-Giacobino A., Rossier C., Dulloo A., Seydoux J., Muzzin P., Giacobino J. P. (1997) FEBS Lett. 408, 39–42 - PubMed
-
- Jaburek M., Varecha M., Gimeno R. E., Dembski M., Jezek P., Zhang M., Burn P., Tartaglia L. A., Garlid K. D. (1999) J. Biol. Chem. 274, 26003–26007 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

