Simultaneous glycan-peptide characterization using hydrophilic interaction chromatography and parallel fragmentation by CID, higher energy collisional dissociation, and electron transfer dissociation MS applied to the N-linked glycoproteome of Campylobacter jejuni
- PMID: 20360033
- PMCID: PMC3033663
- DOI: 10.1074/mcp.M000031-MCP201
Simultaneous glycan-peptide characterization using hydrophilic interaction chromatography and parallel fragmentation by CID, higher energy collisional dissociation, and electron transfer dissociation MS applied to the N-linked glycoproteome of Campylobacter jejuni
Abstract
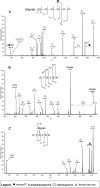
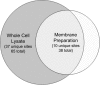
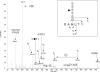
Campylobacter jejuni is a gastrointestinal pathogen that is able to modify membrane and periplasmic proteins by the N-linked addition of a 7-residue glycan at the strict attachment motif (D/E)XNX(S/T). Strategies for a comprehensive analysis of the targets of glycosylation, however, are hampered by the resistance of the glycan-peptide bond to enzymatic digestion or β-elimination and have previously concentrated on soluble glycoproteins compatible with lectin affinity and gel-based approaches. We developed strategies for enriching C. jejuni HB93-13 glycopeptides using zwitterionic hydrophilic interaction chromatography and examined novel fragmentation, including collision-induced dissociation (CID) and higher energy collisional (C-trap) dissociation (HCD) as well as CID/electron transfer dissociation (ETD) mass spectrometry. CID/HCD enabled the identification of glycan structure and peptide backbone, allowing glycopeptide identification, whereas CID/ETD enabled the elucidation of glycosylation sites by maintaining the glycan-peptide linkage. A total of 130 glycopeptides, representing 75 glycosylation sites, were identified from LC-MS/MS using zwitterionic hydrophilic interaction chromatography coupled to CID/HCD and CID/ETD. CID/HCD provided the majority of the identifications (73 sites) compared with ETD (26 sites). We also examined soluble glycoproteins by soybean agglutinin affinity and two-dimensional electrophoresis and identified a further six glycosylation sites. This study more than doubles the number of confirmed N-linked glycosylation sites in C. jejuni and is the first to utilize HCD fragmentation for glycopeptide identification with intact glycan. We also show that hydrophobic integral membrane proteins are significant targets of glycosylation in this organism. Our data demonstrate that peptide-centric approaches coupled to novel mass spectrometric fragmentation techniques may be suitable for application to eukaryotic glycoproteins for simultaneous elucidation of glycan structures and peptide sequence.
Figures






Similar articles
-
Use of CID/ETD mass spectrometry to analyze glycopeptides.Curr Protoc Protein Sci. 2012 Apr;Chapter 12:12.11.1-12.11.11. doi: 10.1002/0471140864.ps1211s68. Curr Protoc Protein Sci. 2012. PMID: 22470127 Free PMC article.
-
Enrichment and identification of bacterial glycopeptides by mass spectrometry.Methods Mol Biol. 2015;1295:355-68. doi: 10.1007/978-1-4939-2550-6_25. Methods Mol Biol. 2015. PMID: 25820733
-
Characterization of intact N- and O-linked glycopeptides using higher energy collisional dissociation.Anal Biochem. 2014 May 1;452:96-102. doi: 10.1016/j.ab.2014.01.003. Epub 2014 Jan 15. Anal Biochem. 2014. PMID: 24440233 Free PMC article.
-
Liquid chromatography-tandem mass spectrometry-based fragmentation analysis of glycopeptides.Glycoconj J. 2016 Jun;33(3):261-72. doi: 10.1007/s10719-016-9649-3. Epub 2016 Jan 18. Glycoconj J. 2016. PMID: 26780731 Review.
-
[Recent advances in glycopeptide enrichment and mass spectrometry data interpretation approaches for glycoproteomics analyses].Se Pu. 2021 Oct;39(10):1045-1054. doi: 10.3724/SP.J.1123.2021.06011. Se Pu. 2021. PMID: 34505426 Free PMC article. Review. Chinese.
Cited by
-
Structure of human ST8SiaIII sialyltransferase provides insight into cell-surface polysialylation.Nat Struct Mol Biol. 2015 Aug;22(8):627-35. doi: 10.1038/nsmb.3060. Epub 2015 Jul 20. Nat Struct Mol Biol. 2015. PMID: 26192331
-
High-sensitivity analytical approaches for the structural characterization of glycoproteins.Chem Rev. 2013 Apr 10;113(4):2668-732. doi: 10.1021/cr3003714. Epub 2013 Mar 27. Chem Rev. 2013. PMID: 23531120 Free PMC article. Review. No abstract available.
-
Mass Spectrometry Targeted Assays as a Tool to Improve Our Understanding of Post-translational Modifications in Pathogenic Bacteria.Front Microbiol. 2016 Aug 4;7:1216. doi: 10.3389/fmicb.2016.01216. eCollection 2016. Front Microbiol. 2016. PMID: 27540373 Free PMC article. No abstract available.
-
Targeted identification of glycosylated proteins in the gastric pathogen Helicobacter pylori (Hp).Mol Cell Proteomics. 2013 Sep;12(9):2568-86. doi: 10.1074/mcp.M113.029561. Epub 2013 Jun 10. Mol Cell Proteomics. 2013. PMID: 23754784 Free PMC article.
-
Separation and identification of isomeric glycopeptides by high field asymmetric waveform ion mobility spectrometry.Anal Chem. 2012 Mar 6;84(5):2597-601. doi: 10.1021/ac203321y. Epub 2012 Feb 10. Anal Chem. 2012. PMID: 22280549 Free PMC article.
References
-
- Zilbauer M., Dorrell N., Wren B. W., Bajaj-Elliott M. (2008) Campylobacter jejuni-mediated disease pathogenesis: an update. Trans. R. Soc. Trop. Med. Hyg. 102, 123–129 - PubMed
-
- Young K. T., Davis L. M., Dirita V. J. (2007) Campylobacter jejuni: molecular biology and pathogenesis. Nat. Rev. Microbiol. 5, 665–679 - PubMed
-
- Nachamkin I. (2002) Chronic effects of Campylobacter infection. Microbes Infect. 4, 399–403 - PubMed
-
- Lecuit M., Abachin E., Martin A., Poyart C., Pochart P., Suarez F., Bengoufa D., Feuillard J., Lavergne A., Gordon J. I., Berche P., Guillevin L., Lortholary O. (2004) Immunoproliferative small intestinal disease associated with Campylobacter jejuni. N. Engl. J. Med. 350, 239–248 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

