Deep mRNA sequencing for in vivo functional analysis of cardiac transcriptional regulators: application to Galphaq
- PMID: 20360248
- PMCID: PMC2891025
- DOI: 10.1161/CIRCRESAHA.110.217513
Deep mRNA sequencing for in vivo functional analysis of cardiac transcriptional regulators: application to Galphaq
Abstract
Rationale: Transcriptional profiling can detect subclinical heart disease and provide insight into disease etiology and functional status. Current microarray-based methods are expensive and subject to artifact.
Objective: To develop RNA sequencing methodologies using next generation massively parallel platforms for high throughput comprehensive analysis of individual mouse cardiac transcriptomes. To compare the results of sequencing- and array-based transcriptional profiling in the well-characterized Galphaq transgenic mouse hypertrophy/cardiomyopathy model.
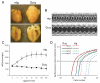
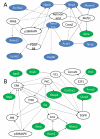
Methods and results: The techniques for preparation of individually bar-coded mouse heart RNA libraries for Illumina Genome Analyzer II resequencing are described. RNA sequencing showed that 234 high-abundance transcripts (>60 copies/cell) comprised 55% of total cardiac mRNA. Parallel transcriptional profiling of Galphaq transgenic and nontransgenic hearts by Illumina RNA sequencing and Affymetrix Mouse Gene 1.0 ST arrays revealed superior dynamic range for mRNA expression and enhanced specificity for reporting low-abundance transcripts by RNA sequencing. Differential mRNA expression in Galphaq and nontransgenic hearts correlated well between microarrays and RNA sequencing for highly abundant transcripts. RNA sequencing was superior to arrays for accurately quantifying lower-abundance genes, which represented the majority of the regulated genes in the Galphaq transgenic model.
Conclusions: RNA sequencing is rapid, accurate, and sensitive for identifying both abundant and rare cardiac transcripts, and has significant advantages in time- and cost-efficiencies over microarray analysis.
Figures


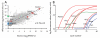
Similar articles
-
Mitochondrial reprogramming induced by CaMKIIδ mediates hypertrophy decompensation.Circ Res. 2015 Feb 27;116(5):e28-39. doi: 10.1161/CIRCRESAHA.116.304682. Epub 2015 Jan 20. Circ Res. 2015. PMID: 25605649 Free PMC article.
-
Nuclear effects of G-protein receptor kinase 5 on histone deacetylase 5-regulated gene transcription in heart failure.Circ Heart Fail. 2011 Sep;4(5):659-68. doi: 10.1161/CIRCHEARTFAILURE.111.962563. Epub 2011 Jul 18. Circ Heart Fail. 2011. PMID: 21768220 Free PMC article.
-
Next-generation sequencing facilitates quantitative analysis of wild-type and Nrl(-/-) retinal transcriptomes.Mol Vis. 2011;17:3034-54. Epub 2011 Nov 23. Mol Vis. 2011. PMID: 22162623 Free PMC article.
-
Physiologic growth and pathologic genes in cardiac development and cardiomyopathy.Trends Cardiovasc Med. 2005 Jul;15(5):185-9. doi: 10.1016/j.tcm.2005.05.009. Trends Cardiovasc Med. 2005. PMID: 16165015 Review.
-
Deep sequencing of coding and non-coding RNA in the CNS.Brain Res. 2010 Jun 18;1338:146-54. doi: 10.1016/j.brainres.2010.03.039. Epub 2010 Mar 20. Brain Res. 2010. PMID: 20307502 Free PMC article. Review.
Cited by
-
Whole genome expression differences in human left and right atria ascertained by RNA sequencing.Circ Cardiovasc Genet. 2012 Jun;5(3):327-35. doi: 10.1161/CIRCGENETICS.111.961631. Epub 2012 Apr 3. Circ Cardiovasc Genet. 2012. PMID: 22474228 Free PMC article.
-
Ovarian transcriptome associated with reproductive senescence in the long-living Ames dwarf mice.Mol Cell Endocrinol. 2017 Jan 5;439:328-336. doi: 10.1016/j.mce.2016.09.019. Epub 2016 Sep 20. Mol Cell Endocrinol. 2017. PMID: 27663076 Free PMC article.
-
A deep sequencing approach to uncover the miRNOME in the human heart.PLoS One. 2013;8(2):e57800. doi: 10.1371/journal.pone.0057800. Epub 2013 Feb 27. PLoS One. 2013. PMID: 23460909 Free PMC article.
-
RISC RNA sequencing for context-specific identification of in vivo microRNA targets.Circ Res. 2011 Jan 7;108(1):18-26. doi: 10.1161/CIRCRESAHA.110.233528. Epub 2010 Oct 28. Circ Res. 2011. PMID: 21030712 Free PMC article.
-
Analysis of transcriptome complexity through RNA sequencing in normal and failing murine hearts.Circ Res. 2011 Dec 9;109(12):1332-41. doi: 10.1161/CIRCRESAHA.111.249433. Epub 2011 Oct 27. Circ Res. 2011. PMID: 22034492 Free PMC article.
References
-
- Dorn GW, II, Robbins J, Sugden PH. Phenotyping hypertrophy - eschew obfuscation. Circ Res. 2003;92:1171–1175. - PubMed
-
- Dorn GW., II The fuzzy logic of physiological cardiac hypertrophy. Hypertension. 2007;49:962–970. - PubMed
-
- Dorn GW, II, Matkovich SJ. Put your chips on transcriptomics. Circulation. 2008;118:216–218. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

