High-resolution profiling of gammaH2AX around DNA double strand breaks in the mammalian genome
- PMID: 20360682
- PMCID: PMC2868577
- DOI: 10.1038/emboj.2010.38
High-resolution profiling of gammaH2AX around DNA double strand breaks in the mammalian genome
Abstract
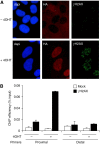
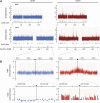
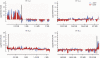
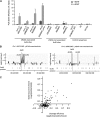
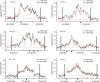
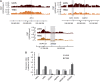
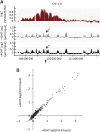
Chromatin acts as a key regulator of DNA-related processes such as DNA damage repair. Although ChIP-chip is a powerful technique to provide high-resolution maps of protein-genome interactions, its use to study DNA double strand break (DSB) repair has been hindered by the limitations of the available damage induction methods. We have developed a human cell line that permits induction of multiple DSBs randomly distributed and unambiguously positioned within the genome. Using this system, we have generated the first genome-wide mapping of gammaH2AX around DSBs. We found that all DSBs trigger large gammaH2AX domains, which spread out from the DSB in a bidirectional, discontinuous and not necessarily symmetrical manner. The distribution of gammaH2AX within domains is influenced by gene transcription, as parallel mappings of RNA Polymerase II and strand-specific expression showed that gammaH2AX does not propagate on active genes. In addition, we showed that transcription is accurately maintained within gammaH2AX domains, indicating that mechanisms may exist to protect gene transcription from gammaH2AX spreading and from the chromatin rearrangements induced by DSBs.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

References
-
- Agger K, Santoni-Rugiu E, Holmberg C, Karlstrom O, Helin K (2005) Conditional E2F1 activation in transgenic mice causes testicular atrophy and dysplasia mimicking human CIS. Oncogene 24: 780–789 - PubMed
-
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K (2007) High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 - PubMed
-
- Berkovich E, Monnat RJ Jr, Kastan MB (2007) Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat Cell Biol 9: 683–690 - PubMed
-
- Celeste A, Fernandez-Capetillo O, Kruhlak MJ, Pilch DR, Staudt DW, Lee A, Bonner RF, Bonner WM, Nussenzweig A (2003) Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat Cell Biol 5: 675–679 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

